Evidence that the Amyloid beta Precursor Protein-intracellular domain lowers the stress threshold of neurons and has a "regulated" transcriptional role
- PMID: 18764939
- PMCID: PMC2538519
- DOI: 10.1186/1750-1326-3-12
Evidence that the Amyloid beta Precursor Protein-intracellular domain lowers the stress threshold of neurons and has a "regulated" transcriptional role
Abstract
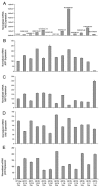
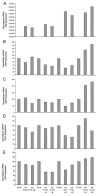
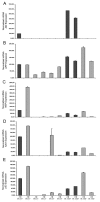
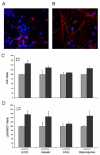
Background: Regulated intramembrane proteolysis of the beta-amyloid precursor protein by the gamma-secretase yields two peptides. One, amyloid-beta, is the major component of the amyloid plaques found in Alzheimer's disease patients. The other, APP IntraCellular Domain, has been involved in regulation of apoptosis, calcium flux and gene transcription. To date, a few potential target genes transcriptionally controlled by AID, alone or complexed with Fe65/Tip60, have been described. Although the reports are controversial: these include KAI1, Neprilysin, p53, EGFR, LRP and APP itself. Furthermore, p53 has been implicated in AID mediated susceptibility to apoptosis. To extend these findings, and assess their in vivo relevance, we have analyzed the expression of the putative target genes and of the total brain basal transriptoma in transgenic mice expressing AID in the forebrain. Also, we have studied the susceptibility of primary neurons from such mice to stress and pro-apoptotic agents.
Results: We found that AID-target genes and the mouse brain basal transcriptoma are not influenced by transgenic expression of AID alone, in the absence of Fe65 over-expression. Also, experiments conducted on primary neurons from AID transgenic mice, suggest a role for AID in sensitizing these cells to toxic stimuli. Overall, these findings hint that a role for AID, in regulating gene transcription, could be induced by yet undefined, and possibly stressful, stimuli in vivo.
Conclusion: Overall, these data suggest that the release of the APP intracellular domain may modulate the sensitivity of neuronal cells to toxic stimuli, and that a transcriptional role of AID could be inscribed in signaling pathways thatare not activated in basal conditions.
Figures

References
-
- Passer B, Pellegrini L, Russo C, Siegel RM, Lenardo MJ, Schettini G, Bachmann M, Tabaton M, D'Adamio L. Generation of an apoptotic intracellular peptide by gamma-secretase cleavage of Alzheimer's amyloid beta protein precursor. J Alzheimers Dis. 2000;2:289–301. - PubMed
-
- Leissring MA, Murphy MP, Mead TR, Akbari Y, Sugarman MC, Jannatipour M, Anliker B, Muller U, Saftig P, De Strooper B, Wolfe MS, Golde TE, LaFerla FM. Proc Natl Acad Sci U S A. 2002/03/28. Vol. 99. 2002. A physiologic signaling role for the gamma -secretase-derived intracellular fragment of APP; pp. 4697–4702. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

