Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression
- PMID: 18765666
- PMCID: PMC2662187
- DOI: 10.1074/jbc.M803512200
Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression
Abstract
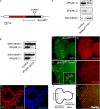
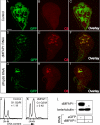
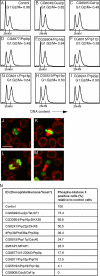
The mammalian spliceosome has mainly been studied using proteomics. The isolation and comparison of different splicing intermediates has revealed the dynamic association of more than 200 splicing factors with the spliceosome, relatively few of which have been studied in detail. Here, we report the characterization of the Drosophila homologue of microfibril-associated protein 1 (dMFAP1), a previously uncharacterized protein found in some human spliceosomal fractions ( Jurica, M. S., and Moore, M. J. (2003) Mol. Cell 12, 5-14 ). We show that dMFAP1 binds directly to the Drosophila homologue of Prp38p (dPrp38), a tri-small nuclear ribonucleoprotein component ( Xie, J., Beickman, K., Otte, E., and Rymond, B. C. (1998) EMBO J. 17, 2938-2946 ), and is required for pre-mRNA processing. dMFAP1, like dPrp38, is essential for viability, and our in vivo data show that cells with reduced levels of dMFAP1 or dPrp38 proliferate more slowly than normal cells and undergo apoptosis. Consistent with this, double-stranded RNA-mediated depletion of dPrp38 or dMFAP1 causes cells to arrest in G(2)/M, and this is paralleled by a reduction in mRNA levels of the mitotic phosphatase string/cdc25. Interestingly double-stranded RNA-mediated depletion of a wide range of core splicing factors elicits a similar phenotype, suggesting that the observed G(2)/M arrest might be a general consequence of interfering with spliceosome function.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

