Models from experiments: combinatorial drug perturbations of cancer cells
- PMID: 18766176
- PMCID: PMC2564730
- DOI: 10.1038/msb.2008.53
Models from experiments: combinatorial drug perturbations of cancer cells
Abstract
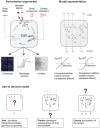
We present a novel method for deriving network models from molecular profiles of perturbed cellular systems. The network models aim to predict quantitative outcomes of combinatorial perturbations, such as drug pair treatments or multiple genetic alterations. Mathematically, we represent the system by a set of nodes, representing molecular concentrations or cellular processes, a perturbation vector and an interaction matrix. After perturbation, the system evolves in time according to differential equations with built-in nonlinearity, similar to Hopfield networks, capable of representing epistasis and saturation effects. For a particular set of experiments, we derive the interaction matrix by minimizing a composite error function, aiming at accuracy of prediction and simplicity of network structure. To evaluate the predictive potential of the method, we performed 21 drug pair treatment experiments in a human breast cancer cell line (MCF7) with observation of phospho-proteins and cell cycle markers. The best derived network model rediscovered known interactions and contained interesting predictions. Possible applications include the discovery of regulatory interactions, the design of targeted combination therapies and the engineering of molecular biological networks.
Figures


References
-
- Chou TC (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58: 621–681 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

