Ultrafast dynamics of flavins in five redox states
- PMID: 18767842
- PMCID: PMC2637805
- DOI: 10.1021/ja8045469
Ultrafast dynamics of flavins in five redox states
Abstract
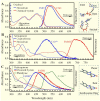
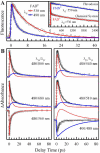
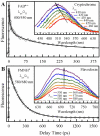
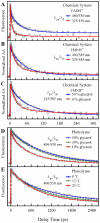
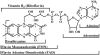
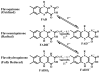
We report here our systematic studies of excited-state dynamics of two common flavin molecules, FMN and FAD, in five redox states--oxidized form, neutral and anionic semiquinones, and neutral and anionic fully reduced hydroquinones--in solution and in inert protein environments with femtosecond resolution. Using protein environments, we were able to stabilize two semiquinone radicals and thus observed their weak emission spectra. Significantly, we observed a strong correlation between their excited-state dynamics and the planarity of their flavin isoalloxazine ring. For a bent ring structure, we observed ultrafast dynamics from a few to hundreds of picoseconds and strong excitation-wavelength dependence of emission spectra, indicating deactivation during relaxation. A butterfly bending motion is invoked to get access to conical intersection(s) to facilitate deactivation. These states include the anionic semiquinone radical and fully reduced neutral and anionic hydroquinones in solution. In a planar configuration, flavins have a long lifetime of nanoseconds, except for the stacked conformation of FAD, where intramolecular electron transfer between the ring and the adenine moiety in 5-9 ps as well as subsequent charge recombination in 30-40 ps were observed. These observed distinct dynamics, controlled by the flavin ring flexibility, are fundamental to flavoenzyme's functions, as observed in photolyase with a planar structure to lengthen the lifetime to maximize DNA repair efficiency and in insect type 1 cryptochrome with a flexible structure to vary the excited-state deactivation to modulate the functional channel.
Figures



References
-
- Walsh C. Acc. Chem. Res. 1980;13:148–155.
-
- Massey V, Hemmerich P. Biochem. Soc. Trans. 1980;8:246–257. - PubMed
-
- Müller F. The Flavin Redox-System and Its Biological Function. In: Boschke FL, editor. Topics in Current Chemistry. Vol. 108. Springer -Verlag; Berlin: 1983. pp. 71–107. - PubMed
-
- Müller F, editor. Chemistry and Biochemistry of Flavoenzymes. I-III. CRC Press; Boca Raton, FL: 19901991.
-
- Fraaije MW, Mattevi A. Trends Biochem. Sci. 2000;25:126–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

