Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates
- PMID: 18768804
- PMCID: PMC2533215
- DOI: 10.1073/pnas.0802426105
Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates
Abstract
Vertebrate ancestors appeared in a uniform, shallow water environment, but modern species flourish in highly variable niches. A striking array of phenotypes exhibited by contemporary animals is assumed to have evolved by accumulating a series of selectively advantageous mutations. However, the experimental test of such adaptive events at the molecular level is remarkably difficult. One testable phenotype, dim-light vision, is mediated by rhodopsins. Here, we engineered 11 ancestral rhodopsins and show that those in early ancestors absorbed light maximally (lambda(max)) at 500 nm, from which contemporary rhodopsins with variable lambda(max)s of 480-525 nm evolved on at least 18 separate occasions. These highly environment-specific adaptations seem to have occurred largely by amino acid replacements at 12 sites, and most of those at the remaining 191 ( approximately 94%) sites have undergone neutral evolution. The comparison between these results and those inferred by commonly-used parsimony and Bayesian methods demonstrates that statistical tests of positive selection can be misleading without experimental support and that the molecular basis of spectral tuning in rhodopsins should be elucidated by mutagenesis analyses using ancestral pigments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

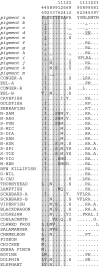
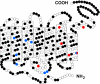
Comment in
-
The origin of adaptive phenotypes.Proc Natl Acad Sci U S A. 2008 Sep 9;105(36):13193-4. doi: 10.1073/pnas.0807440105. Epub 2008 Sep 3. Proc Natl Acad Sci U S A. 2008. PMID: 18768803 Free PMC article. No abstract available.
References
-
- Lewontin RC. Adaptation. Sci Amer. 1978;239:3–13. - PubMed
-
- Yokoyama S. Molecular genetic basis of adaptive selection: Examples from colour vision in vertebrates. Annu Rev Genet. 1977;31:315–336. - PubMed
-
- Yang Z. PAML 4: A program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. - PubMed
-
- Suzuki Y, Gojobori T, Nei M. ADAPTSITE: Detecting natural selection at single nucleotide sites. Bioinformatics. 2001;17:660–661. - PubMed
-
- Zhang J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet. 2006;38:819–823. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

