A catalog of neutral and deleterious polymorphism in yeast
- PMID: 18769710
- PMCID: PMC2515631
- DOI: 10.1371/journal.pgen.1000183
A catalog of neutral and deleterious polymorphism in yeast
Abstract
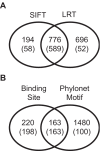
The abundance and identity of functional variation segregating in natural populations is paramount to dissecting the molecular basis of quantitative traits as well as human genetic diseases. Genome sequencing of multiple organisms of the same species provides an efficient means of cataloging rearrangements, insertion, or deletion polymorphisms (InDels) and single-nucleotide polymorphisms (SNPs). While inbreeding depression and heterosis imply that a substantial amount of polymorphism is deleterious, distinguishing deleterious from neutral polymorphism remains a significant challenge. To identify deleterious and neutral DNA sequence variation within Saccharomyces cerevisiae, we sequenced the genome of a vineyard and oak tree strain and compared them to a reference genome. Among these three strains, 6% of the genome is variable, mostly attributable to variation in genome content that results from large InDels. Out of the 88,000 polymorphisms identified, 93% are SNPs and a small but significant fraction can be attributed to recent interspecific introgression and ectopic gene conversion. In comparison to the reference genome, there is substantial evidence for functional variation in gene content and structure that results from large InDels, frame-shifts, and polymorphic start and stop codons. Comparison of polymorphism to divergence reveals scant evidence for positive selection but an abundance of evidence for deleterious SNPs. We estimate that 12% of coding and 7% of noncoding SNPs are deleterious. Based on divergence among 11 yeast species, we identified 1,666 nonsynonymous SNPs that disrupt conserved amino acids and 1,863 noncoding SNPs that disrupt conserved noncoding motifs. The deleterious coding SNPs include those known to affect quantitative traits, and a subset of the deleterious noncoding SNPs occurs in the promoters of genes that show allele-specific expression, implying that some cis-regulatory SNPs are deleterious. Our results show that the genome sequences of both closely and distantly related species provide a means of identifying deleterious polymorphisms that disrupt functionally conserved coding and noncoding sequences.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Hypervariable noncoding sequences in Saccharomyces cerevisiae.Genetics. 2005 Aug;170(4):1575-87. doi: 10.1534/genetics.105.042283. Epub 2005 Jun 14. Genetics. 2005. PMID: 15956668 Free PMC article.
-
Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species.Genome Res. 2010 Nov;20(11):1558-73. doi: 10.1101/gr.108993.110. Epub 2010 Sep 4. Genome Res. 2010. PMID: 20817943 Free PMC article.
-
A unique ecological niche fosters hybridization of oak-tree and vineyard isolates of Saccharomyces cerevisiae.Mol Ecol. 2015 Dec;24(23):5886-98. doi: 10.1111/mec.13439. Epub 2015 Nov 20. Mol Ecol. 2015. PMID: 26518477 Free PMC article.
-
Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes.Toxicol Appl Pharmacol. 2005 Sep 1;207(2 Suppl):84-90. doi: 10.1016/j.taap.2004.09.024. Toxicol Appl Pharmacol. 2005. PMID: 16002116 Review.
-
Methods to detect selection on noncoding DNA.Methods Mol Biol. 2012;856:141-59. doi: 10.1007/978-1-61779-585-5_6. Methods Mol Biol. 2012. PMID: 22399458 Free PMC article. Review.
Cited by
-
Initiating-clone analysis in patients with acute myeloid leukemia secondary to essential thrombocythemia.Sci Rep. 2024 Jul 10;14(1):15906. doi: 10.1038/s41598-024-66461-8. Sci Rep. 2024. PMID: 38987297 Free PMC article.
-
Computational analysis of high-risk SNPs in human CHK2 gene responsible for hereditary breast cancer: A functional and structural impact.PLoS One. 2019 Aug 9;14(8):e0220711. doi: 10.1371/journal.pone.0220711. eCollection 2019. PLoS One. 2019. PMID: 31398194 Free PMC article.
-
Prediction of Deleterious Non-synonymous SNPs of Human STK11 Gene by Combining Algorithms, Molecular Docking, and Molecular Dynamics Simulation.Sci Rep. 2019 Nov 11;9(1):16426. doi: 10.1038/s41598-019-52308-0. Sci Rep. 2019. PMID: 31712642 Free PMC article.
-
Idiosyncratic epistasis leads to global fitness-correlated trends.Science. 2022 May 6;376(6593):630-635. doi: 10.1126/science.abm4774. Epub 2022 May 5. Science. 2022. PMID: 35511982 Free PMC article.
-
Bioinformatics classification of mutations in patients with Mucopolysaccharidosis IIIA.Metab Brain Dis. 2019 Dec;34(6):1577-1594. doi: 10.1007/s11011-019-00465-6. Epub 2019 Aug 5. Metab Brain Dis. 2019. PMID: 31385193 Free PMC article.
References
-
- Lewontin R. The genetic basis of evolutionary change. New York: Columbia University Press; 1974. 374
-
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. - PubMed
-
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander E. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

