Binomial mitotic segregation of MYCN-carrying double minutes in neuroblastoma illustrates the role of randomness in oncogene amplification
- PMID: 18769732
- PMCID: PMC2518122
- DOI: 10.1371/journal.pone.0003099
Binomial mitotic segregation of MYCN-carrying double minutes in neuroblastoma illustrates the role of randomness in oncogene amplification
Abstract
Background: Amplification of the oncogene MYCN in double minutes (DMs) is a common finding in neuroblastoma (NB). Because DMs lack centromeric sequences it has been unclear how NB cells retain and amplify extrachromosomal MYCN copies during tumour development.
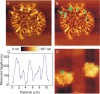
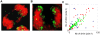
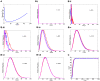
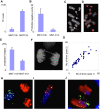
Principal findings: We show that MYCN-carrying DMs in NB cells translocate from the nuclear interior to the periphery of the condensing chromatin at transition from interphase to prophase and are preferentially located adjacent to the telomere repeat sequences of the chromosomes throughout cell division. However, DM segregation was not affected by disruption of the telosome nucleoprotein complex and DMs readily migrated from human to murine chromatin in human/mouse cell hybrids, indicating that they do not bind to specific positional elements in human chromosomes. Scoring DM copy-numbers in ana/telophase cells revealed that DM segregation could be closely approximated by a binomial random distribution. Colony-forming assay demonstrated a strong growth-advantage for NB cells with high DM (MYCN) copy-numbers, compared to NB cells with lower copy-numbers. In fact, the overall distribution of DMs in growing NB cell populations could be readily reproduced by a mathematical model assuming binomial segregation at cell division combined with a proliferative advantage for cells with high DM copy-numbers.
Conclusion: Binomial segregation at cell division explains the high degree of MYCN copy-number variability in NB. Our findings also provide a proof-of-principle for oncogene amplification through creation of genetic diversity by random events followed by Darwinian selection.
Conflict of interest statement
Figures



References
-
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. - PubMed
-
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. - PubMed
-
- Norris MD, Bordow SB, Haber PS, Marshall GM, Kavallaris M, et al. Evidence that the MYCN oncogene regulates MRP gene expression in neuroblastoma. Eur J Cancer. 1997;33:1911–1916. - PubMed
-
- Shohet JM, Hicks MJ, Plon SE, Burlingame SM, Stuart S, et al. Minichromosome maintenance protein MCM7 is a direct target of the MYCN transcription factor in neuroblastoma. Cancer Res. 2002;62:1123–1128. - PubMed
-
- Narath R, Ambros IM, Kowalska A, Bozsaky E, Boukamp P, et al. Induction of senescence in MYCN amplified neuroblastoma cell lines by hydroxyurea. Genes Chromosomes Cancer. 2007;46:130–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

