Regulation of the mouse Treacher Collins syndrome homolog (Tcof1) promoter through differential repression of constitutive expression
- PMID: 18771418
- PMCID: PMC2925028
- DOI: 10.1089/dna.2008.0766
Regulation of the mouse Treacher Collins syndrome homolog (Tcof1) promoter through differential repression of constitutive expression
Abstract
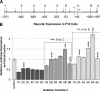
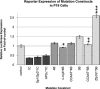
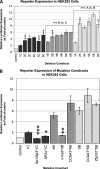
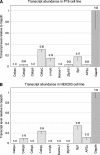
Treacher Collins syndrome is an autosomal-dominant mandibulofacial dysostosis caused by haploinsufficiency of the TCOF1 gene product treacle. Mouse Tcof1 protein is approximately 61% identical and 71% similar to treacle, and heterozygous knockout of Tcof1 causes craniofacial malformation. Tcof1 expression is high in developing neural crest, but much lower in other tissues. To investigate this dual regulation, highly conserved regions upstream of TCOF1 homologs were tested through deletion and mutation reporter assays, and conserved predicted transcription factor binding sites were assessed through chromatin binding studies. Assays were performed in mouse P19 embryonic carcinoma cells and in HEK293 cells to determine differential activation in cell types at different stages of differentiation. Binding of Cebpb, Zfp161, and Sp1 transcription factors was specific to the Tcof1 regulatory region in P19 cells. The Zfp161 binding site demonstrated P19 cell-specific repression, while the Sp1/Sp3 candidate site demonstrated HEK293 cell-specific activation. Moreover, presence of c-myb and Zfp161 transcripts was specific to P19 cells. A minimal promoter fragment from -253 to +43 bp directs constitutive expression in both cell types, and dual regulation of Tcof1 appears to be through differential repression of this minimal promoter. The CpG island at the transcription start site remains unmethylated in P19 cells, 11.5 dpc mouse embryonic tissue, and adult mouse ear, which supports constitutive activation of the Tcof1 promoter.
Figures



References
-
- Borchers A. David R. Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. - PubMed
-
- Dixon J. Dixon M.J. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein Treacle. Dev Dyn. 2004;229:907–914. - PubMed
-
- Dixon J. Edwards S.J. Anderson I. Brass A. Scambler P.J. Dixon M.J. Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res. 1997a;7:223–234. - PubMed
-
- Dixon J. Hovanes K. Shiang R. Dixon M.J. Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Hum Mol Genet. 1997b;6:727–737. - PubMed
-
- Dixon J. Brakebusch C. Fassler R. Dixon M.J. Increased levels of apoptosis in the prefusion neural folds underlie the craniofacial disorder, Treacher Collins syndrome. Hum Mol Genet. 2000;9:1473–1480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

