DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome
- PMID: 18771760
- PMCID: PMC2556438
- DOI: 10.1016/j.ajhg.2008.08.012
DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome
Abstract
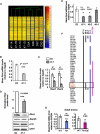
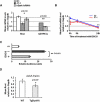
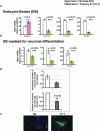
Down syndrome (DS) is the most common cause of mental retardation. Many neural phenotypes are shared between DS individuals and DS mouse models; however, the common underlying molecular pathogenetic mechanisms remain unclear. Using a transchromosomic model of DS, we show that a 30%-60% reduced expression of Nrsf/Rest (a key regulator of pluripotency and neuronal differentiation) is an alteration that persists in trisomy 21 from undifferentiated embryonic stem (ES) cells to adult brain and is reproducible across several DS models. Using partially trisomic ES cells, we map this effect to a three-gene segment of HSA21, containing DYRK1A. We independently identify the same locus as the most significant eQTL controlling REST expression in the human genome. We show that specifically silencing the third copy of DYRK1A rescues Rest levels, and we demonstrate altered Rest expression in response to inhibition of DYRK1A expression or kinase activity, and in a transgenic Dyrk1A mouse. We reveal that undifferentiated trisomy 21 ES cells show DYRK1A-dose-sensitive reductions in levels of some pluripotency regulators, causing premature expression of transcription factors driving early endodermal and mesodermal differentiation, partially overlapping recently reported downstream effects of Rest +/-. They produce embryoid bodies with elevated levels of the primitive endoderm progenitor marker Gata4 and a strongly reduced neuroectodermal progenitor compartment. Our results suggest that DYRK1A-mediated deregulation of REST is a very early pathological consequence of trisomy 21 with potential to disturb the development of all embryonic lineages, warranting closer research into its contribution to DS pathology and new rationales for therapeutic approaches.
Figures


References
-
- Epstein C. Down Syndrome. In: Scriver C.R.B.A., Sly W.S., Valle D., editors. The metabolic and molecular bases of inherited disease. McGraw-Hill; New York: 2001. pp. 1223–1256.
-
- Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 2004;5:725–738. - PubMed
-
- Reeves R.H., Irving N.G., Moran T.H., Wohn A., Kitt C., Sisodia S.S., Schmidt C., Bronson R.T., Davisson M.T. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 1995;11:177–184. - PubMed
-
- Smith D.J., Stevens M.E., Sudanagunta S.P., Bronson R.T., Makhinson M., Watabe A.M., O'Dell T.J., Fung J., Weier H.U., Cheng J.F. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 1997;16:28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

