Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge
- PMID: 18772133
- PMCID: PMC2581559
- DOI: 10.1074/jbc.M804828200
Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge
Abstract
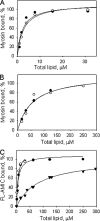
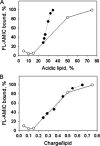
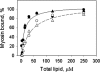
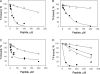
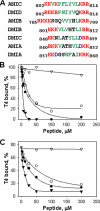
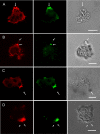
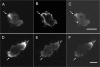
The tail of Acanthamoeba myosin IC (AMIC) has a basic region (BR), which contains a putative pleckstrin homology (PH) domain, followed by two Gly/Pro/Ala (GPA)-rich regions separated by a Src homology 3 (SH3) domain. Cryoelectron microscopy had shown that the tail is folded back on itself at the junction of BR and GPA1, and nuclear magnetic resonance spectroscopy indicated that the SH3 domain may interact with the putative PH domain. The BR binds to acidic phospholipids, and the GPA region binds to F-actin. We now show that the folded tail does not affect the affinity of AMIC for acidic phospholipids. AMIC binds phosphatidylinositol 4,5-bisphosphate (PIP2) with high affinity (approximately 1 microm), but binding is not stereospecific. When normalized to net negative charge, AMIC binds with equal affinity to phosphatidylserine (PS) and PIP2. This and other data show that the putative PH domain of AMIC is not a typical PIP2-specific PH domain. We have identified a 13-residue sequence of basic-hydrophobic-basic amino acids within the putative PH domain that may be a major determinant of binding of AMIC to acidic phospholipids. Despite the lack of stereospecificity, AMIC binds 10 times more strongly to vesicles containing 5% PIP2 plus 25% PS than to vesicles containing only 25% PS, suggesting that AMIC may be targeted to PIP2-enriched regions of the plasma membrane. In agreement with this, AMIC colocalizes with PIP2 at dynamic, protrusive regions of the plasma membrane. We discuss the possibility that AMIC binding to PIP2 may initiate the formation of a multiprotein complex at the plasma membrane.
Figures

References
-
- Pollard, T. D., Doberstein, S. K., and Zot, H. G. (1991) Annu. Rev. Physiol. 53 653-681 - PubMed
-
- Sellers, J. R. (2000) Biochim. Biophys. Acta 1496 3-22 - PubMed
-
- Brzeska, H., Lynch, T. J., and Korn, E. D. (1988) J. Biol. Chem. 263 427-435 - PubMed
-
- Lynch, T. J., Albanesi, J. P., Korn, E. D., Robinson, E. A., Bowers, B., and Fujisaki, H. (1986) J. Biol. Chem. 261 17156-17162 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

