Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin
- PMID: 18772227
- PMCID: PMC2553572
- DOI: 10.1093/nar/gkn550
Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin
Abstract
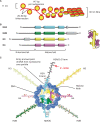
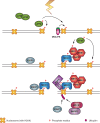
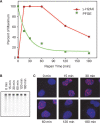
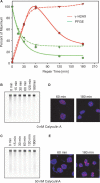
DNA double-strand breaks (DSBs) are extremely dangerous lesions with severe consequences for cell survival and the maintenance of genomic stability. In higher eukaryotic cells, DSBs in chromatin promptly initiate the phosphorylation of the histone H2A variant, H2AX, at Serine 139 to generate gamma-H2AX. This phosphorylation event requires the activation of the phosphatidylinositol-3-OH-kinase-like family of protein kinases, DNA-PKcs, ATM, and ATR, and serves as a landing pad for the accumulation and retention of the central components of the signaling cascade initiated by DNA damage. Regions in chromatin with gamma-H2AX are conveniently detected by immunofluorescence microscopy and serve as beacons of DSBs. This has allowed the development of an assay that has proved particularly useful in the molecular analysis of the processing of DSBs. Here, we first review the role of gamma-H2AX in DNA damage response in the context of chromatin and discuss subsequently the use of this modification as a surrogate marker for mechanistic studies of DSB induction and processing. We conclude with a critical analysis of the strengths and weaknesses of the approach and present some interesting applications of the resulting methodology.
Figures



References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond S, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 2004;1677:3–11. - PubMed
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–668. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

