Cross-species virus transmission and the emergence of new epidemic diseases
- PMID: 18772285
- PMCID: PMC2546865
- DOI: 10.1128/MMBR.00004-08
Cross-species virus transmission and the emergence of new epidemic diseases
Abstract
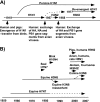
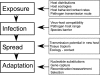
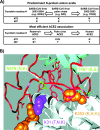
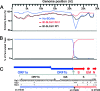
Host range is a viral property reflecting natural hosts that are infected either as part of a principal transmission cycle or, less commonly, as "spillover" infections into alternative hosts. Rarely, viruses gain the ability to spread efficiently within a new host that was not previously exposed or susceptible. These transfers involve either increased exposure or the acquisition of variations that allow them to overcome barriers to infection of the new hosts. In these cases, devastating outbreaks can result. Steps involved in transfers of viruses to new hosts include contact between the virus and the host, infection of an initial individual leading to amplification and an outbreak, and the generation within the original or new host of viral variants that have the ability to spread efficiently between individuals in populations of the new host. Here we review what is known about host switching leading to viral emergence from known examples, considering the evolutionary mechanisms, virus-host interactions, host range barriers to infection, and processes that allow efficient host-to-host transmission in the new host population.
Figures



References
-
- Anonymous. December 2007, posting date. AIDS epidemic update. http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf. UNAIDS, Geneva, Switzerland.
-
- Bauch, C. T., J. O. Lloyd-Smith, M. P. Coffee, and A. P. Galvani. 2005. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology 16791-801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

