Comprehensive genomic characterization defines human glioblastoma genes and core pathways
- PMID: 18772890
- PMCID: PMC2671642
- DOI: 10.1038/nature07385
Comprehensive genomic characterization defines human glioblastoma genes and core pathways
Erratum in
- Nature. 2013 Feb 28;494(7438):506
Abstract
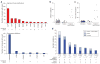
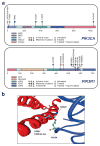
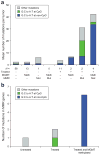
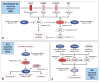
Human cancer cells typically harbour multiple chromosomal aberrations, nucleotide substitutions and epigenetic modifications that drive malignant transformation. The Cancer Genome Atlas (TCGA) pilot project aims to assess the value of large-scale multi-dimensional analysis of these molecular characteristics in human cancer and to provide the data rapidly to the research community. Here we report the interim integrative analysis of DNA copy number, gene expression and DNA methylation aberrations in 206 glioblastomas--the most common type of adult brain cancer--and nucleotide sequence aberrations in 91 of the 206 glioblastomas. This analysis provides new insights into the roles of ERBB2, NF1 and TP53, uncovers frequent mutations of the phosphatidylinositol-3-OH kinase regulatory subunit gene PIK3R1, and provides a network view of the pathways altered in the development of glioblastoma. Furthermore, integration of mutation, DNA methylation and clinical treatment data reveals a link between MGMT promoter methylation and a hypermutator phenotype consequent to mismatch repair deficiency in treated glioblastomas, an observation with potential clinical implications. Together, these findings establish the feasibility and power of TCGA, demonstrating that it can rapidly expand knowledge of the molecular basis of cancer.
Figures

References
-
- Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. - PubMed
-
- Mischel PS, Nelson SF, Cloughesy TF. Molecular analysis of glioblastoma: pathway profiling and its implications for patient therapy. Cancer Biol Ther. 2003;2:242–7. - PubMed
-
- Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. - PubMed
-
- Maher EA, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24CA126561/CA/NCI NIH HHS/United States
- U24CA126551/CA/NCI NIH HHS/United States
- R01 CA099041/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA126546/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- U24CA126543/CA/NCI NIH HHS/United States
- U24CA126554/CA/NCI NIH HHS/United States
- U24CA126544/CA/NCI NIH HHS/United States
- U24 CA126551/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA126554/CA/NCI NIH HHS/United States
- K08 NS045077/NS/NINDS NIH HHS/United States
- U24 CA126561/CA/NCI NIH HHS/United States
- U24 CA126543/CA/NCI NIH HHS/United States
- U24CA126563/CA/NCI NIH HHS/United States
- U24CA126546/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24 CA126563/CA/NCI NIH HHS/United States
- U24 CA126544/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

