NRAGE, a p75NTR adaptor protein, is required for developmental apoptosis in vivo
- PMID: 18772898
- PMCID: PMC2735073
- DOI: 10.1038/cdd.2008.127
NRAGE, a p75NTR adaptor protein, is required for developmental apoptosis in vivo
Abstract
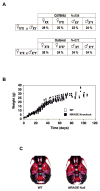
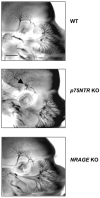
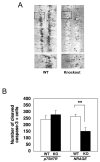
NRAGE (also known as Maged1, Dlxin) is a member of the MAGE gene family that may play a role in the neuronal apoptosis that is regulated by the p75 neurotrophin receptor (p75NTR). To test this hypothesis in vivo, we generated NRAGE knockout mice and found that NRAGE deletion caused a defect in developmental apoptosis of sympathetic neurons of the superior cervical ganglia, similar to that observed in p75NTR knockout mice. Primary sympathetic neurons derived from NRAGE knockout mice were resistant to apoptosis induced by brain-derived neurotrophic factor (BDNF), a pro-apoptotic p75NTR ligand, and NRAGE-deficient sympathetic neurons show attenuated BDNF-dependent JNK activation. Hair follicle catagen is an apoptosis-like process that is dependent on p75NTR signaling; we show that NRAGE and p75NTR show regulated co-expression in the hair follicle and that identical defects in hair follicle catagen are present in NRAGE and p75NTR knockout mice. Interestingly, NRAGE knockout mice have severe defects in motoneuron apoptosis that are not observed in p75NTR knockout animals, raising the possibility that NRAGE may facilitate apoptosis induced by receptors other than p75NTR. Together, these studies demonstrate that NRAGE plays an important role in apoptotic-signaling in vivo.
Figures


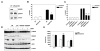
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

