Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish
- PMID: 18773071
- PMCID: PMC2518861
- DOI: 10.1371/journal.pgen.1000174
Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish
Abstract
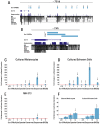
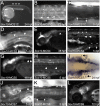
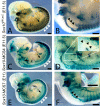
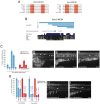
Sox10 is a dynamically regulated transcription factor gene that is essential for the development of neural crest-derived and oligodendroglial populations. Developmental genes often require multiple regulatory sequences that integrate discrete and overlapping functions to coordinate their expression. To identify Sox10 cis-regulatory elements, we integrated multiple model systems, including cell-based screens and transposon-mediated transgensis in zebrafish, to scrutinize mammalian conserved, noncoding genomic segments at the mouse Sox10 locus. We demonstrate that eight of 11 Sox10 genomic elements direct reporter gene expression in transgenic zebrafish similar to patterns observed in transgenic mice, despite an absence of observable sequence conservation between mice and zebrafish. Multiple segments direct expression in overlapping populations of neural crest derivatives and glial cells, ranging from pan-Sox10 and pan-neural crest regulatory control to the modulation of expression in subpopulations of Sox10-expressing cells, including developing melanocytes and Schwann cells. Several sequences demonstrate overlapping spatial control, yet direct expression in incompletely overlapping developmental intervals. We were able to partially explain neural crest expression patterns by the presence of head to head SoxE family binding sites within two of the elements. Moreover, we were able to use this transcription factor binding site signature to identify the corresponding zebrafish enhancers in the absence of overall sequence homology. We demonstrate the utility of zebrafish transgenesis as a high-fidelity surrogate in the dissection of mammalian gene regulation, especially those with dynamically controlled developmental expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kalcheim C, Le Douarin NM. Requirement of a neural tube signal for the differentiation of neural crest cells into dorsal root ganglia. Dev Biol. 1986;116:451–466. - PubMed
-
- Bondurand N, Kobetz A, Pingault V, Lemort N, Encha-Razavi F, et al. Expression of the SOX10 gene during human development. FEBS Lett. 1998;432:168–172. - PubMed
-
- Pusch C, Hustert E, Pfeifer D, Sudbeck P, Kist R, et al. The SOX10/Sox10 gene from human and mouse: sequence, expression, and transactivation by the encoded HMG domain transcription factor. Hum Genet. 1998;103:115–123. - PubMed
-
- Kapur RP. Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatr Dev Pathol. 1999;2:559–569. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

