A Sox10 expression screen identifies an amino acid essential for Erbb3 function
- PMID: 18773073
- PMCID: PMC2518866
- DOI: 10.1371/journal.pgen.1000177
A Sox10 expression screen identifies an amino acid essential for Erbb3 function
Abstract
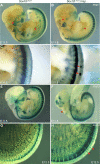
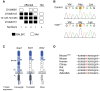
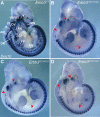
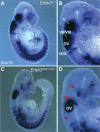
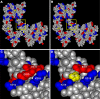
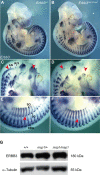
The neural crest (NC) is a population of embryonic stem cells that gives rise to numerous cell types, including the glia and neurons of the peripheral and enteric nervous systems and the melanocytes of the skin and hair. Mutations in genes and genetic pathways regulating NC development lead to a wide spectrum of human developmental disorders collectively called neurocristopathies. To identify molecular pathways regulating NC development and to understand how alterations in these processes lead to disease, we established an N-ethyl-N-nitrosourea (ENU) mutagenesis screen utilizing a mouse model sensitized for NC defects, Sox10(LacZ/+). Out of 71 pedigrees analyzed, we identified and mapped four heritable loci, called modifier of Sox10 expression pattern 1-4 (msp1-4), which show altered NC patterning. In homozygous msp1 embryos, Sox10(LacZ) expression is absent in cranial ganglia, cranial nerves, and the sympathetic chain; however, the development of other Sox10-expressing cells appears unaffected by the mutation. Linkage analysis, sequencing, and complementation testing confirmed that msp1 is a new allele of the receptor tyrosine kinase Erbb3, Erbb3(msp1), that carries a single amino acid substitution in the extracellular region of the protein. The ENU-induced mutation does not alter protein expression, however, it is sufficient to impair ERBB3 signaling such that the embryonic defects observed in msp1 resemble those of Erbb3 null alleles. Biochemical analysis of the mutant protein showed that ERBB3 is expressed on the cell surface, but its ligand-induced phosphorylation is dramatically reduced by the msp1 mutation. These findings highlight the importance of the mutated residue for ERBB3 receptor function and activation. This study underscores the utility of using an ENU mutagenesis to identify genetic pathways regulating NC development and to dissect the roles of discrete protein domains, both of which contribute to a better understanding of gene function in a cellular and developmental setting.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

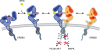
References
-
- Newgreen DF, Erickson CA. The migration of neural crest cells. Int Rev Cytol. 1986;103:89–145. - PubMed
-
- Loring JF, Erickson CA. Neural crest cell migratory pathways in the trunk of the chick embryo. Dev Biol. 1987;121:220–236. - PubMed
-
- Le Douarin NM, Kalcheim C. Cambridge, UK: The Neural Crest, Second Edition Cambridge University Press; 1999.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

