Coupling endoplasmic reticulum stress to the cell death program in dopaminergic cells: effect of paraquat
- PMID: 18773310
- PMCID: PMC2818083
- DOI: 10.1007/s12017-008-8047-9
Coupling endoplasmic reticulum stress to the cell death program in dopaminergic cells: effect of paraquat
Abstract
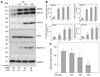
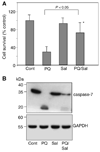
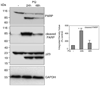
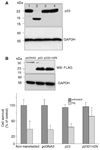
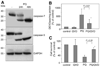
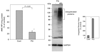
Parkinson's disease (PD) features oxidative stress and accumulation of misfolded (unfolded, alternatively folded, or mutant) proteins with associated loss of dopaminergic neurons. Oxidative stress and the accumulated misfolded proteins elicit cellular responses that include an endoplasmic reticulum (ER) stress response that may protect cells against the toxic buildup of misfolded proteins. Chronic ER stress and accumulation of misfolded proteins in excessive amounts, however, overwhelm the cellular 'quality control' system and impair the protective mechanisms designed to promote correct folding and degrade faulty proteins, ultimately leading to organelle dysfunction and neuronal cell death. Paraquat belongs to a class of bipyridyl herbicides and triggers oxidative stress and dopaminergic cell death. Epidemiological studies suggest an increased risk for developing PD following chronic exposure to paraquat. The present study was carried out to determine the role of paraquat in triggering cellular stress particularly ER stress and to elucidate the pathways that couple ER stress to dopaminergic cell death. We demonstrate that paraquat triggers ER stress, cell dysfunction, and dopaminergic cell death. p23, a small co-chaperone protein, is cleaved during ER stress-induced cell death triggered by paraquat and blockage of the caspase cleavage site of p23 was associated with decreased cell death. Paraquat also inhibits proteasomal activity that may further trigger accumulation of misfolded proteins resulting in ER stress. Our results indicate a protective role for p23 in PD-related programmed cell death. The data also underscore the involvement of ER, caspases, and the proteasomal system in ER stress-induced cell death process.
Figures
References
-
- Adams FS, La Rosa FG, Kumar S, Edwards-Prasad J, Kentroti S, Vernadakis A, et al. Characterization and transplantation of two neuronal cell lines with dopaminergic properties. Neurochemical Research. 1996;21:619–627. doi: 10.1007/BF02527762. - DOI - PubMed
-
- Andersen JK. What causes the build-up of ubiquitin-containing inclusions in Parkinson’s disease? Mechanisms of Ageing and Development. 2000;118:15–22. doi: 10.1016/S0047-6374 (00)00150-0. - DOI - PubMed
-
- Andersen JK. Paraquat and iron exposure as possible synergistic environmental risk factors in Parkinson’s disease. Neurotoxic Research. 2003;5:307–313. - PubMed
-
- Andersen JK. Oxidative stress in neurodegeneration: Cause or consequence? Nature Medicine. 2004;10 Suppl:S18–S25. doi: 10.1038/nrn1434. - DOI - PubMed
-
- Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

