Cis-active RNA elements (CREs) and picornavirus RNA replication
- PMID: 18773930
- PMCID: PMC2692539
- DOI: 10.1016/j.virusres.2008.07.027
Cis-active RNA elements (CREs) and picornavirus RNA replication
Abstract
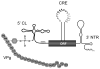
Our understanding of picornavirus RNA replication has improved over the past 10 years, due in large part to the discovery of cis-active RNA elements (CREs) within picornavirus RNA genomes. CREs function as templates for the conversion of VPg, the Viral Protein of the genome, into VPgpUpU(OH). These so called CREs are different from the previously recognized cis-active RNA sequences and structures within the 5' and 3' NTRs of picornavirus genomes. Two adenosine residues in the loop of the CRE RNA structures allow the viral RNA-dependent RNA polymerase 3D(Pol) to add two uridine residues to the tyrosine residue of VPg. Because VPg and/or VPgpUpU(OH) prime the initiation of viral RNA replication, the asymmetric replication of viral RNA could not be explained without an understanding of the viral RNA template involved in the conversion of VPg into VPgpUpU(OH) primers. We review the growing body of knowledge regarding picornavirus CREs and discuss how CRE RNAs work coordinately with viral replication proteins and other cis-active RNAs in the 5' and 3' NTRs during RNA replication.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

