The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining
- PMID: 18773976
- PMCID: PMC2644637
- DOI: 10.1016/j.dnarep.2008.08.002
The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining
Abstract
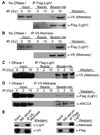
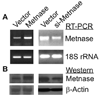
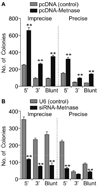
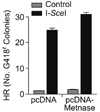
Transposase domain proteins mediate DNA movement from one location in the genome to another in lower organisms. However, in human cells such DNA mobility would be deleterious, and therefore the vast majority of transposase-related sequences in humans are pseudogenes. We recently isolated and characterized a SET and transposase domain protein termed Metnase that promotes DNA double-strand break (DSB) repair by non-homologous end-joining (NHEJ). Both the SET and transposase domain were required for its NHEJ activity. In this study we found that Metnase interacts with DNA Ligase IV, an important component of the classical NHEJ pathway. We investigated whether Metnase had structural requirements of the free DNA ends for NHEJ repair, and found that Metnase assists in joining all types of free DNA ends equally well. Metnase also prevents long deletions from processing of the free DNA ends, and improves the accuracy of NHEJ. Metnase levels correlate with the speed of disappearance of gamma-H2Ax sites after ionizing radiation. However, Metnase has little effect on homologous recombination repair of a single DSB. Altogether, these results fit a model where Metnase plays a role in the fate of free DNA ends during NHEJ repair of DSBs.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. - PubMed
-
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. - PubMed
-
- Fu YP, Yu JC, Cheng TC, Lou MA, Hsu GC, Wu CY, Chen ST, Wu HS, Wu PE, Shen CY. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. - PubMed
-
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

