Antioxidants, HIF prolyl hydroxylase inhibitors or short interfering RNAs to BNIP3 or PUMA, can prevent prodeath effects of the transcriptional activator, HIF-1alpha, in a mouse hippocampal neuronal line
- PMID: 18774900
- PMCID: PMC2612757
- DOI: 10.1089/ars.2008.2039
Antioxidants, HIF prolyl hydroxylase inhibitors or short interfering RNAs to BNIP3 or PUMA, can prevent prodeath effects of the transcriptional activator, HIF-1alpha, in a mouse hippocampal neuronal line
Abstract
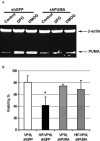
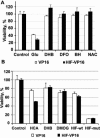
Hypoxia-inducible factor (HIF) is a transcriptional activator that promotes death or survival in neurons. The regulators and targets of HIF-1alpha-mediated death remain unclear. We found that prodeath effects of HIF-1 are not attributable to an imbalance in HIF-1alpha and HIF-1beta expression. Rather, the synergistic death caused by oxidative stress and by overexpression of an oxygen-resistant HIF-VP16 in neuroblasts was attributable to transcriptional upregulation of BH3-only prodeath proteins, PUMA or BNIP3. By contrast, overexpression of BNIP3 was not sufficient to potentiate oxidative death. As acidosis is known to activate BNIP3-mediated death, we examined other secondary stresses, such as oxidants or prolyl hydroxylase activity are necessary for exposing the prodeath functions of HIF in neurons. Antioxidants or prolyl hydroxylase inhibition prevented potentiation of death by HIF-1alpha. Together, these studies suggest that antioxidants and PHD inhibitors abrogate the ability of HIF-mediated transactivation of BH3-only proteins to potentiate oxidative death in normoxia. The findings offer strategies for minimizing the prodeath effects of HIF-1 in neurologic conditions associated with hypoxia and oxidative stress, such as stroke and spinal cord injury.
Figures







References
-
- Aminova LR. Chavez JC. Lee J. Ryu H. Kung A. Lamanna JC. Ratan RR. Prosurvival, prodeath effects of hypoxia-inducible factor-1alpha stabilization in a murine hippocampal cell line. J Biol Chem. 2005;280:3996–4003. - PubMed
-
- Chen D. Li M. Luo J. Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278:13595–13598. - PubMed
-
- Conkright MD. Guzman E. Flechner L. Su AI. Hogenesch JB. Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11:1101–1108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

