Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia
- PMID: 18775315
- PMCID: PMC2570342
- DOI: 10.1016/j.cell.2008.06.054
Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia
Abstract
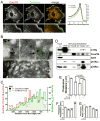
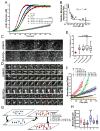
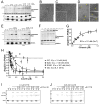
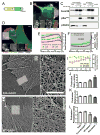
The dendritic actin network generated by the Arp2/3 complex in lamellipodia underlies formation of protrusions, directional sensing, and migration. While the generation of this network is well studied, the mechanisms regulating network disassembly are poorly understood. We report that Coronin 1B disassembles Arp2/3-containing actin filament branches by inducing Arp2/3 dissociation. This activity is antagonized by Cortactin, a filament branch stabilizer. Consistent with this biochemical competition, depletion of both proteins partially rescues defects in lamellipodial dynamics observed upon depletion of either protein alone. Coronin 1B targets actin branches in a manner that is mutually exclusive with the Arp2/3 complex and alters the branch angle. We conclude that Coronin 1B replaces the Arp2/3 complex at actin filament branches as the dendritic network matures and drives the turnover of branched actin networks.
Figures


Similar articles
-
Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge.Cell. 2007 Mar 9;128(5):915-29. doi: 10.1016/j.cell.2007.01.031. Cell. 2007. PMID: 17350576 Free PMC article.
-
Cortactin stabilizes actin branches by bridging activated Arp2/3 to its nucleated actin filament.Nat Struct Mol Biol. 2024 May;31(5):801-809. doi: 10.1038/s41594-023-01205-2. Epub 2024 Jan 24. Nat Struct Mol Biol. 2024. PMID: 38267598 Free PMC article.
-
Isoform diversity in the Arp2/3 complex determines actin filament dynamics.Nat Cell Biol. 2016 Jan;18(1):76-86. doi: 10.1038/ncb3286. Epub 2015 Dec 14. Nat Cell Biol. 2016. PMID: 26655834
-
Coronin: the double-edged sword of actin dynamics.Subcell Biochem. 2008;48:72-87. doi: 10.1007/978-0-387-09595-0_7. Subcell Biochem. 2008. PMID: 18925372 Review.
-
Cortactin: a multifunctional regulator of cellular invasiveness.Cell Adh Migr. 2011 Mar-Apr;5(2):187-98. doi: 10.4161/cam.5.2.14773. Epub 2011 Mar 1. Cell Adh Migr. 2011. PMID: 21258212 Free PMC article. Review.
Cited by
-
Lamellipodia are crucial for haptotactic sensing and response.J Cell Sci. 2016 Jun 15;129(12):2329-42. doi: 10.1242/jcs.184507. Epub 2016 May 12. J Cell Sci. 2016. PMID: 27173494 Free PMC article.
-
Macropinocytosis and Cell Migration: Don't Drink and Drive….Subcell Biochem. 2022;98:85-102. doi: 10.1007/978-3-030-94004-1_5. Subcell Biochem. 2022. PMID: 35378704
-
Cellular and pathophysiological consequences of Arp2/3 complex inhibition: role of inhibitory proteins and pharmacological compounds.Cell Mol Life Sci. 2019 Sep;76(17):3349-3361. doi: 10.1007/s00018-019-03128-y. Epub 2019 May 9. Cell Mol Life Sci. 2019. PMID: 31073744 Free PMC article. Review.
-
Bi-modal regulation of a formin by srGAP2.J Biol Chem. 2011 Feb 25;286(8):6577-86. doi: 10.1074/jbc.M110.190397. Epub 2010 Dec 9. J Biol Chem. 2011. PMID: 21148482 Free PMC article.
-
PDK1-mediated activation of MRCKα regulates directional cell migration and lamellipodia retraction.J Cell Biol. 2014 Aug 4;206(3):415-34. doi: 10.1083/jcb.201312090. J Cell Biol. 2014. PMID: 25092657 Free PMC article.
References
-
- Anes E, Kuhnel MP, Bos E, Moniz-Pereira J, Habermann A, Griffiths G. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat Cell Biol. 2003;5:793–802. - PubMed
-
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. - PubMed
-
- Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. - PubMed
-
- Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J Biol Chem. 2005;280:31913–31923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

