Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing
- PMID: 18775325
- PMCID: PMC2696193
- DOI: 10.1016/j.molcel.2008.06.020
Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing
Abstract
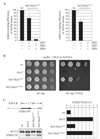
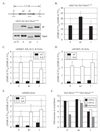
The yeast Sir2/3/4 complex forms a heterochromatin-like structure that represses transcription. The proteins nucleate at silencers and spread distally, utilizing the Sir2 NAD(+)-dependent histone deacetylase activity and the affinity of Sir3/4 for deacetylated histone tails. A by-product of the Sir2 reaction, O-acetyl-ADP-ribose (OAADPr), is thought to aid spreading by binding one of the Sir proteins. We developed a protein chimera approach to reexamine the contributions of Sir2. We show that a Sir3 chimera-bearing Hos3, an unrelated NAD(+)-independent histone deacetylase, substitutes for Sir2 in silencing. Sir3-Hos3 operates within the Sir pathway, spreading while deacetylating histones. Moreover, the chimera represses HM loci in strains lacking all five OAADPr-producing deacetylases, indicating that OAADPr is not necessary for silencing. Repression by a Hos3 hybrid bearing the targeting motifs of Sir2 shows that targeting doesn't require the Sir2 reaction. Together, these data demonstrate that protein deacetylation is the only essential function of Sir2 in creating silenced chromatin.
Figures





References
-
- Ahn SH, Diaz RL, Grunstein M, Allis CD. Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol. Cell. 2006;24:211–220. - PubMed
-
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SirT1. J. Biol. Chem. 2002;277:45099–45107. - PubMed
-
- Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 2002;277:12632–12641. - PubMed
-
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a 'silencer' in yeast: a DNA sequence with properties opposite those of a transcriptional enhancer. Cell. 1985;41:41–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

