Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity
- PMID: 18775691
- PMCID: PMC2655699
- DOI: 10.1016/j.ydbio.2008.08.013
Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity
Abstract
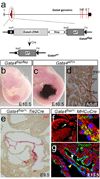
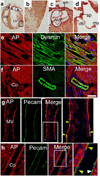
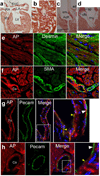
Isl1 and Nkx2-5-expressing cardiovascular progenitors play pivotal roles in cardiogenesis. Previously reported Cre-based fate-mapping studies showed that Isl1 progenitors contribute predominantly to the derivatives of the second heart field, and Nkx2-5 progenitors contributed mainly to the cardiomyocyte lineage. However, partial recombination of Cre reporter genes can complicate interpretation of Cre fate-mapping experiments. We found that a Gata4-based Cre-activated reporter was recombined by Isl1(Cre) and Nkx2-5(Cre) in a substantially broader domain than previously reported using standard Cre-activated reporters. The expanded Isl1 and Nkx2-5 cardiac fate maps were remarkably similar, and included extensive contributions to cardiomyocyte, endocardial, and smooth muscle lineages in all four cardiac chambers. These data indicate that Isl1 is expressed in progenitors of both primary and secondary heart fields, and that Nkx2-5 is expressed in progenitors of cardiac endothelium and smooth muscle, in addition to cardiomyocytes. These results have important implications for our understanding of cardiac lineage diversification in vivo, and for the interpretation of Cre-based fate maps.
Figures



References
-
- Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–285. - PubMed
-
- Brade T, Gessert S, Kuhl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007 - PubMed
-
- Bruneau BG, Black BL. The heart's Da Vinci code: a Renaissance at Keystone. Development. 2007;134:1631–1633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

