Rapid whole-genome mutational profiling using next-generation sequencing technologies
- PMID: 18775913
- PMCID: PMC2556265
- DOI: 10.1101/gr.077776.108
Rapid whole-genome mutational profiling using next-generation sequencing technologies
Abstract
Forward genetic mutational studies, adaptive evolution, and phenotypic screening are powerful tools for creating new variant organisms with desirable traits. However, mutations generated in the process cannot be easily identified with traditional genetic tools. We show that new high-throughput, massively parallel sequencing technologies can completely and accurately characterize a mutant genome relative to a previously sequenced parental (reference) strain. We studied a mutant strain of Pichia stipitis, a yeast capable of converting xylose to ethanol. This unusually efficient mutant strain was developed through repeated rounds of chemical mutagenesis, strain selection, transformation, and genetic manipulation over a period of seven years. We resequenced this strain on three different sequencing platforms. Surprisingly, we found fewer than a dozen mutations in open reading frames. All three sequencing technologies were able to identify each single nucleotide mutation given at least 10-15-fold nominal sequence coverage. Our results show that detecting mutations in evolved and engineered organisms is rapid and cost-effective at the whole-genome level using new sequencing technologies. Identification of specific mutations in strains with altered phenotypes will add insight into specific gene functions and guide further metabolic engineering efforts.
Figures
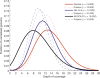
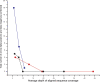
References
-
- Burke D., Dawson D., Stearns T., Dawson D., Stearns T., Stearns T., et al. Cold Spring Hargor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. Methods in yeast genetics.
-
- Gordon D., Abajian C., Green P., Abajian C., Green P., Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. - PubMed
-
- Jeffries T.W., Grigoriev I.V., Grimwood J., Laplaza J.M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Grigoriev I.V., Grimwood J., Laplaza J.M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Grimwood J., Laplaza J.M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Laplaza J.M., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Aerts A., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Salamov A., Schmutz J., Lindquist E., Dehal P., Shapiro H., Schmutz J., Lindquist E., Dehal P., Shapiro H., Lindquist E., Dehal P., Shapiro H., Dehal P., Shapiro H., Shapiro H., et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007;25:319–326. - PubMed
-
- Marth G.T., Korf I., Yandell M.D., Yeh R.T., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Korf I., Yandell M.D., Yeh R.T., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Yandell M.D., Yeh R.T., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Yeh R.T., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Gu Z., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Zakeri H., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Stitziel N.O., Hillier L., Kwok P.Y., Gish W.R., Hillier L., Kwok P.Y., Gish W.R., Kwok P.Y., Gish W.R., Gish W.R. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 1999;23:452–456. - PubMed
-
- Parekh S.R., Parekh R.S., Wayman M., Parekh R.S., Wayman M., Wayman M. Fermentation of xylose and cellobiose by Pichia stipitis and Brettanomyces clausenii. Appl. Biochem. Biotechnol. 1988;18:325–338.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
