Copy number variation and evolution in humans and chimpanzees
- PMID: 18775914
- PMCID: PMC2577862
- DOI: 10.1101/gr.082016.108
Copy number variation and evolution in humans and chimpanzees
Abstract
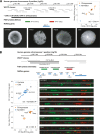
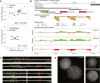
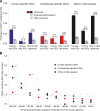
Copy number variants (CNVs) underlie many aspects of human phenotypic diversity and provide the raw material for gene duplication and gene family expansion. However, our understanding of their evolutionary significance remains limited. We performed comparative genomic hybridization on a single human microarray platform to identify CNVs among the genomes of 30 humans and 30 chimpanzees as well as fixed copy number differences between species. We found that human and chimpanzee CNVs occur in orthologous genomic regions far more often than expected by chance and are strongly associated with the presence of highly homologous intrachromosomal segmental duplications. By adapting population genetic analyses for use with copy number data, we identified functional categories of genes that have likely evolved under purifying or positive selection for copy number changes. In particular, duplications and deletions of genes with inflammatory response and cell proliferation functions may have been fixed by positive selection and involved in the adaptive phenotypic differentiation of humans and chimpanzees.
Figures


References
-
- Akashi H. Within- and between-species DNA sequence variation and the 'footprint' of natural selection. Gene. 1999;238:39–51. - PubMed
-
- Asthana S., Schmidt S., Sunyaev S. A limited role for balancing selection. Trends Genet. 2005;21:30–32. - PubMed
-
- Babcock M., Yatsenko S., Hopkins J., Brenton M., Cao Q., de Jong P., Stankiewicz P., Lupski J.R., Sikela J.M., Morrow B.E. Hominoid lineage specific amplification of low-copy repeats on 22q11.2 (LCR22s) associated with velo-cardio-facial/digeorge syndrome. Hum. Mol. Genet. 2007;16:2560–2571. - PubMed
-
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
