Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways
- PMID: 18775984
- PMCID: PMC2581544
- DOI: 10.1074/jbc.M806127200
Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways
Abstract
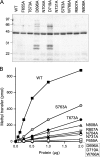
Trimethylguanosine synthase (Tgs1) is the enzyme that converts standard m(7)G caps to the 2,2,7-trimethylguanosine (TMG) caps characteristic of spliceosomal small nuclear RNAs. Fungi and mammalian somatic cells are able to grow in the absence of Tgs1 and TMG caps, suggesting that an essential function of the TMG cap might be obscured by functional redundancy. A systematic screen in budding yeast identified nonessential genes that, when deleted, caused synthetic growth defects with tgs1Delta. The Tgs1 interaction network embraced proteins implicated in small nuclear ribonucleoprotein function and spliceosome assembly, including Mud2, Nam8, Brr1, Lea1, Ist3, Isy1, Cwc21, and Bud13. Complementation of the synthetic lethality of mud2Delta tgs1Delta and nam8Delta tgs1Delta strains by wild-type TGS1, but not by catalytically defective mutants, indicated that the TMG cap is essential for mitotic growth when redundant splicing factors are missing. Our genetic analysis also highlighted synthetic interactions of Tgs1 with proteins implicated in RNA end processing and decay (Pat1, Lsm1, and Trf4) and regulation of polymerase II transcription (Rpn4, Spt3, Srb2, Soh1, Swr1, and Htz1). We find that the C-terminal domain of human Tgs1 can function in lieu of the yeast protein in vivo. We present a biochemical characterization of the human Tgs1 guanine-N2 methyltransferase reaction and identify individual amino acids required for methyltransferase activity in vitro and in vivo.
Figures





Similar articles
-
Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing.RNA. 2010 May;16(5):1018-31. doi: 10.1261/rna.2082610. Epub 2010 Apr 1. RNA. 2010. PMID: 20360394 Free PMC article.
-
Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1Δ cells.Nucleic Acids Res. 2011 Aug;39(15):6715-28. doi: 10.1093/nar/gkr279. Epub 2011 May 10. Nucleic Acids Res. 2011. PMID: 21558325 Free PMC article.
-
An essential role for trimethylguanosine RNA caps in Saccharomyces cerevisiae meiosis and their requirement for splicing of SAE3 and PCH2 meiotic pre-mRNAs.Nucleic Acids Res. 2011 Jul;39(13):5633-46. doi: 10.1093/nar/gkr083. Epub 2011 Mar 11. Nucleic Acids Res. 2011. PMID: 21398639 Free PMC article.
-
Two Routes to Genetic Suppression of RNA Trimethylguanosine Cap Deficiency via C-Terminal Truncation of U1 snRNP Subunit Snp1 or Overexpression of RNA Polymerase Subunit Rpo26.G3 (Bethesda). 2015 Apr 24;5(7):1361-70. doi: 10.1534/g3.115.016675. G3 (Bethesda). 2015. PMID: 25911228 Free PMC article.
-
Anomalous HIV-1 RNA, How Cap-Methylation Segregates Viral Transcripts by Form and Function.Viruses. 2022 Apr 29;14(5):935. doi: 10.3390/v14050935. Viruses. 2022. PMID: 35632676 Free PMC article. Review.
Cited by
-
Structure-function analysis and genetic interactions of the Luc7 subunit of the Saccharomyces cerevisiae U1 snRNP.RNA. 2016 Sep;22(9):1302-10. doi: 10.1261/rna.056911.116. Epub 2016 Jun 27. RNA. 2016. PMID: 27354704 Free PMC article.
-
A novel protein-protein interaction in the RES (REtention and Splicing) complex.J Biol Chem. 2014 Oct 10;289(41):28640-50. doi: 10.1074/jbc.M114.592311. Epub 2014 Aug 26. J Biol Chem. 2014. PMID: 25160624 Free PMC article.
-
Mutational analyses of trimethylguanosine synthase (Tgs1) and Mud2: proteins implicated in pre-mRNA splicing.RNA. 2010 May;16(5):1018-31. doi: 10.1261/rna.2082610. Epub 2010 Apr 1. RNA. 2010. PMID: 20360394 Free PMC article.
-
Combining Chemical Synthesis and Enzymatic Methylation to Access Short RNAs with Various 5' Caps.Chembiochem. 2019 Jul 1;20(13):1693-1700. doi: 10.1002/cbic.201900037. Epub 2019 May 27. Chembiochem. 2019. PMID: 30768827 Free PMC article.
-
A targeted bypass screen identifies Ynl187p, Prp42p, Snu71p, and Cbp80p for stable U1 snRNP/Pre-mRNA interaction.Mol Cell Biol. 2009 Jul;29(14):3941-52. doi: 10.1128/MCB.00384-09. Epub 2009 May 18. Mol Cell Biol. 2009. PMID: 19451230 Free PMC article.
References
-
- Shuman, S. (2001) Cold Spring Harbor Symp. Quant. Biol. 66 301-312 - PubMed
-
- Srinivasan, P., Piano, F., and Shatkin, A. J. (2003) J. Biol. Chem. 278 14168-14173 - PubMed
-
- Takagi, T., Walker, A. K., Sawa, C., Diehn, F., Takase, Y., Blackwell, T. K., and Buratowski, S. (2003) J. Biol. Chem. 278 14174-14184 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

