Lactate and amino acid catabolism in the cheese-ripening yeast Yarrowia lipolytica
- PMID: 18776032
- PMCID: PMC2576680
- DOI: 10.1128/AEM.01519-08
Lactate and amino acid catabolism in the cheese-ripening yeast Yarrowia lipolytica
Abstract
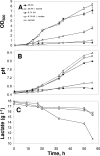
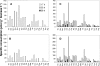
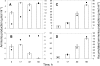
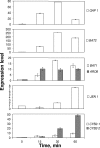
The consumption of lactate and amino acids is very important for microbial development and/or aroma production during cheese ripening. A strain of Yarrowia lipolytica isolated from cheese was grown in a liquid medium containing lactate in the presence of a low (0.1x) or high (2x) concentration of amino acids. Our results show that there was a dramatic increase in the growth of Y. lipolytica in the medium containing a high amino acid concentration, but there was limited lactate consumption. Conversely, lactate was efficiently consumed in the medium containing a low concentration of amino acids after amino acid depletion was complete. These data suggest that the amino acids are used by Y. lipolytica as a main energy source, whereas lactate is consumed following amino acid depletion. Amino acid degradation was accompanied by ammonia production corresponding to a dramatic increase in the pH. The effect of adding amino acids to a Y. lipolytica culture grown on lactate was also investigated. Real-time quantitative PCR analyses were performed with specific primers for five genes involved in amino acid transport and catabolism, including an amino acid transporter gene (GAP1) and four aminotransferase genes (ARO8, ARO9, BAT1, and BAT2). The expression of three genes involved in lactate transport and catabolism was also studied. These genes included a lactate transporter gene (JEN1) and two lactate dehydrogenase genes (CYB2-1 and CYB2-2). Our data showed that GAP1, BAT2, BAT1, and ARO8 were maximally expressed after 15 to 30 min following addition of amino acids (BAT2 was the most highly expressed gene), while the maximum expression of JEN1, CYB2-1, and CYB2-2 was delayed (>or=60 min).
Figures
Similar articles
-
Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis.Appl Environ Microbiol. 2009 Oct;75(20):6422-30. doi: 10.1128/AEM.00228-09. Epub 2009 Aug 14. Appl Environ Microbiol. 2009. PMID: 19684166 Free PMC article.
-
Transcriptomic response of Debaryomyces hansenii during mixed culture in a liquid model cheese medium with Yarrowia lipolytica.Int J Food Microbiol. 2018 Jan 2;264:53-62. doi: 10.1016/j.ijfoodmicro.2017.10.026. Epub 2017 Oct 24. Int J Food Microbiol. 2018. PMID: 29111498
-
A proteomic and transcriptomic view of amino acids catabolism in the yeast Yarrowia lipolytica.Proteomics. 2009 Oct;9(20):4714-25. doi: 10.1002/pmic.200900161. Proteomics. 2009. PMID: 19764064
-
Cheese yeasts.Yeast. 2019 Mar;36(3):129-141. doi: 10.1002/yea.3368. Epub 2019 Jan 24. Yeast. 2019. PMID: 30512214 Review.
-
The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications.Biotechnol Adv. 2011 Nov-Dec;29(6):632-44. doi: 10.1016/j.biotechadv.2011.04.005. Epub 2011 Apr 28. Biotechnol Adv. 2011. PMID: 21550394 Review.
Cited by
-
Engineering the Yeast Yarrowia lipolytica for Production of Polylactic Acid Homopolymer.Front Bioeng Biotechnol. 2020 Oct 22;8:954. doi: 10.3389/fbioe.2020.00954. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33195110 Free PMC article.
-
Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis.Appl Environ Microbiol. 2009 Oct;75(20):6422-30. doi: 10.1128/AEM.00228-09. Epub 2009 Aug 14. Appl Environ Microbiol. 2009. PMID: 19684166 Free PMC article.
-
Microbiological and biochemical aspects of inland Pecorino Abruzzese cheese.Heliyon. 2017 Feb 28;3(2):e00258. doi: 10.1016/j.heliyon.2017.e00258. eCollection 2017 Feb. Heliyon. 2017. PMID: 28280791 Free PMC article.
-
Global regulation of the response to sulfur availability in the cheese-related bacterium Brevibacterium aurantiacum.Appl Environ Microbiol. 2011 Feb;77(4):1449-59. doi: 10.1128/AEM.01708-10. Epub 2010 Dec 17. Appl Environ Microbiol. 2011. PMID: 21169450 Free PMC article.
-
New insights into sulfur metabolism in yeasts as revealed by studies of Yarrowia lipolytica.Appl Environ Microbiol. 2013 Feb;79(4):1200-11. doi: 10.1128/AEM.03259-12. Epub 2012 Dec 7. Appl Environ Microbiol. 2013. PMID: 23220962 Free PMC article.
References
-
- Arfi, K., M. N. Leclercq-Perlat, H. E. Spinnler, and P. Bonnarme. 2005. Importance of curd-neutralising yeasts on the aromatic potential of Brevibacterium linens during cheese ripening. Int. Dairy J. 15:883-891.
-
- Barnett, J. A., R. W. Payne, and D. Yarrow. 2000. Yeasts: characteristics and identification. Cambridge University Press, Cambridge, United Kingdom.
-
- Blanchin-Roland, S., G. D. Costa, and C. Gaillardin. 2005. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology 151:3627-3637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

