Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS
- PMID: 18776048
- PMCID: PMC2531065
- DOI: 10.1073/pnas.0806158105
Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS
Abstract
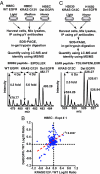
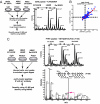
We have used unbiased phosphoproteomic approaches, based on quantitative mass spectrometry using stable isotope labeling with amino acids in cell culture (SILAC), to identify tyrosine phosphorylated proteins in isogenic human bronchial epithelial cells (HBECs) and human lung adenocarcinoma cell lines, expressing either of the two mutant alleles of EGFR (L858R and Del E746-A750), or a mutant KRAS allele, which are common in human lung adenocarcinomas. Tyrosine phosphorylation of signaling molecules was greater in HBECs expressing the mutant EGFRs than in cells expressing WT EGFR or mutant KRAS. Receptor tyrosine kinases (such as EGFR, ERBB2, MET, and IGF1R), and Mig-6, an inhibitor of EGFR signaling, were more phosphorylated in HBECs expressing mutant EGFR than in cells expressing WT EGFR or mutant RAS. Phosphorylation of some proteins differed in the two EGFR mutant-expressing cells; for example, some cell junction proteins (beta-catenin, plakoglobin, and E-cadherin) were more phosphorylated in HBECs expressing L858R EGFR than in cells expressing Del EGFR. There were also differences in degree of phosphorylation at individual tyrosine sites within a protein; for example, a previously uncharacterized phosphorylation site in the nucleotide-binding loop of the kinase domains of EGFR (Y727), ERBB2 (Y735), or ERBB4 (Y733), is phosphorylated significantly more in HBECs expressing the deletion mutant than in cells expressing the wild type or L858R EGFR. Signaling molecules not previously implicated in ERBB signaling, such as polymerase transcript release factor (PTRF), were also phosphorylated in cells expressing mutant EGFR. Bayesian network analysis of these and other datasets revealed that PTRF might be a potentially important component of the ERBB signaling network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
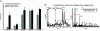


References
-
- Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
-
- Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

