Molecular imaging of proteins in tissues by mass spectrometry
- PMID: 18776051
- PMCID: PMC2587620
- DOI: 10.1073/pnas.0801374105
Molecular imaging of proteins in tissues by mass spectrometry
Abstract
Imaging MS (IMS) is an emerging technology that permits the direct analysis and determination of the distribution of molecules in tissue sections. Biological molecules such as proteins, peptides, lipids, xenobiotics, and metabolites can be analyzed in a high-throughput manner with molecular specificity not readily achievable through other means. Tissues are analyzed intact and thus spatial localization of molecules within a tissue is preserved. Several studies are presented that focus on the unique types of information obtainable by IMS, such as Abeta isoform distributions in Alzheimer's plaques, protein maps in mouse brain, and spatial protein distributions in human breast carcinoma. The analysis of a biopsy taken 100 years ago from a patient with amyloidosis illustrates the use of IMS with formalin-fixed tissues. Finally, the registration and correlation of IMS with MRI is presented.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
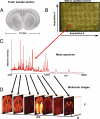
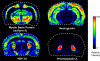
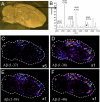
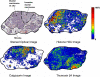
References
-
- McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. - PubMed
-
- Pacholski ML, Winograd N. Imaging with mass spectrometry. Chem Rev. 1999;99:2977–3005. - PubMed
-
- Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption ionization mass spectrometry of biopolymers. Anal Chem. 1991;63:A1193–A1202. - PubMed
-
- Chaurand P, Hayn G, Matter U, Caprioli RM. Exploring the potential of cryodetectors for the detection of matrix-assisted laser desorption/ionization produced ions: Application to profiling and imaging mass spectrometry. Zhipu Xuebao. 2004;25:205–206. 215.
-
- Sugiura Y, Shimma S, Setou M. Thin sectioning improves the peak intensity and signal-to-noise ratio in direct tissue mass spectrometry. J Mass Spectrom Soc Jpn. 2006;54:45–48.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

