RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination
- PMID: 18776220
- PMCID: PMC2566892
- DOI: 10.1093/nar/gkn553
RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination
Abstract
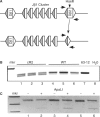
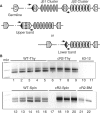
Variable (diversity) joining [V(D)J] recombination of immune gene loci proceeds in an ordered manner with D to J portions recombining first and then an upstream V joins that recombinant. We present evidence that the non-core domain of recombination activating gene (RAG) protein 2 is involved in the regulation of recombinatorial order. In mice lacking the non-core domain of RAG2 the ordered rearrangement is disturbed and direct V to D rearrangements are 10- to 1000-times increased in tri-partite immune gene loci. Some forms of inter-chromosomal translocations between TCRbeta and TCRdelta D gene segments are also increased in the core RAG2 animals as compared with their wild-type (WT) counterparts. In addition, the concise use of proper recombination signal sequences (RSSs) appears to be disturbed in the core RAG2 mice as compared with WT RAG2 animals.
Figures






References
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. - PubMed
-
- VanDyk LF, Wise TW, Moore BB, Meek K. Immunoglobulin D(H) recombination signal sequence targeting: effect of D(H) coding and flanking regions and recombination partner. J. Immunol. 1996;157:4005–4015. - PubMed
-
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. - PubMed

