Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells
- PMID: 18776904
- PMCID: PMC2805063
- DOI: 10.1038/ni.1647
Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells
Abstract
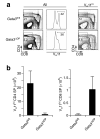
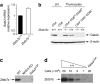
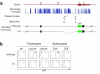
The transcription factors GATA-3 and ThPOK are required for intrathymic differentiation of CD4(+) T cells, but their precise functions in this process remain unclear. Here we show that, contrary to previous findings, Gata3 disruption blocked differentiation into the CD4(+) T cell lineage before commitment to the CD4(+) lineage and in some contexts permitted the 'redirection' of major histocompatibility complex class II-restricted thymocytes into the CD8(+) lineage. GATA-3 promoted ThPOK expression and bound to a region of the locus encoding ThPOK established as being critical for ThPOK expression. Finally, ThPOK promoted differentiation into the CD4(+) lineage in a way dependent on GATA-3 but inhibited differentiation into the CD8(+) lineage independently of GATA-3. We propose that GATA-3 acts as a specification factor for the CD4(+) lineage 'upstream' of the ThPOK-controlled CD4(+) commitment checkpoint.
Figures










Comment in
-
Decision by committee: new light on the CD4/CD8-lineage choice.Immunol Cell Biol. 2009 Feb;87(2):109-12. doi: 10.1038/icb.2008.100. Epub 2008 Dec 16. Immunol Cell Biol. 2009. PMID: 19079359 No abstract available.
References
-
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. - PubMed
-
- Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–540. - PubMed
-
- Laky K, Fowlkes B. Receptor signals and nuclear events in CD4 and CD8 T cell lineage commitment. Curr Opin Immunol. 2005;17:116–121. - PubMed
-
- He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. - PubMed
-
- Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

