A robust statistical method for case-control association testing with copy number variation
- PMID: 18776912
- PMCID: PMC2784596
- DOI: 10.1038/ng.206
A robust statistical method for case-control association testing with copy number variation
Abstract
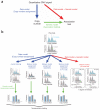
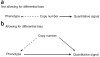
Copy number variation (CNV) is pervasive in the human genome and can play a causal role in genetic diseases. The functional impact of CNV cannot be fully captured through linkage disequilibrium with SNPs. These observations motivate the development of statistical methods for performing direct CNV association studies. We show through simulation that current tests for CNV association are prone to false-positive associations in the presence of differential errors between cases and controls, especially if quantitative CNV measurements are noisy. We present a statistical framework for performing case-control CNV association studies that applies likelihood ratio testing of quantitative CNV measurements in cases and controls. We show that our methods are robust to differential errors and noisy data and can achieve maximal theoretical power. We illustrate the power of these methods for testing for association with binary and quantitative traits, and have made this software available as the R package CNVtools.
Figures
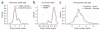
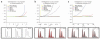
Similar articles
-
Integrated detection and population-genetic analysis of SNPs and copy number variation.Nat Genet. 2008 Oct;40(10):1166-74. doi: 10.1038/ng.238. Epub 2008 Sep 7. Nat Genet. 2008. PMID: 18776908
-
Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs.Nat Genet. 2008 Oct;40(10):1253-60. doi: 10.1038/ng.237. Epub 2008 Sep 7. Nat Genet. 2008. PMID: 18776909 Free PMC article.
-
Systematic assessment of copy number variant detection via genome-wide SNP genotyping.Nat Genet. 2008 Oct;40(10):1199-203. doi: 10.1038/ng.236. Epub 2008 Sep 7. Nat Genet. 2008. PMID: 18776910 Free PMC article.
-
CNV discovery using SNP genotyping arrays.Cytogenet Genome Res. 2008;123(1-4):307-12. doi: 10.1159/000184722. Epub 2009 Mar 11. Cytogenet Genome Res. 2008. PMID: 19287169 Review.
-
Methods and strategies for analyzing copy number variation using DNA microarrays.Nat Genet. 2007 Jul;39(7 Suppl):S16-21. doi: 10.1038/ng2028. Nat Genet. 2007. PMID: 17597776 Free PMC article. Review.
Cited by
-
The Growing Importance of CNVs: New Insights for Detection and Clinical Interpretation.Front Genet. 2013 May 30;4:92. doi: 10.3389/fgene.2013.00092. eCollection 2013. Front Genet. 2013. PMID: 23750167 Free PMC article.
-
Genetic Association Analysis of Copy Number Variations for Meat Quality in Beef Cattle.Foods. 2023 Oct 31;12(21):3986. doi: 10.3390/foods12213986. Foods. 2023. PMID: 37959106 Free PMC article.
-
Utilizing extended pedigree information for discovery and confirmation of copy number variable regions among Mexican Americans.Eur J Hum Genet. 2013 Apr;21(4):404-9. doi: 10.1038/ejhg.2012.188. Epub 2012 Aug 22. Eur J Hum Genet. 2013. PMID: 22909773 Free PMC article.
-
CNVineta: a data mining tool for large case-control copy number variation datasets.Bioinformatics. 2010 Sep 1;26(17):2208-9. doi: 10.1093/bioinformatics/btq356. Epub 2010 Jul 6. Bioinformatics. 2010. PMID: 20605930 Free PMC article.
-
Novel association strategy with copy number variation for identifying new risk Loci of human diseases.PLoS One. 2010 Aug 20;5(8):e12185. doi: 10.1371/journal.pone.0012185. PLoS One. 2010. PMID: 20808825 Free PMC article.
References
-
- Tuzun E, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. - PubMed
-
- Lupski JR, Stankiewicz P, editors. Genomic Disorders: The Genomic Basis of Disease. Humana Press; Totowa, New Jersey: 2006. - PubMed
-
- Flint J, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

