CCL3L1-CCR5 genotype improves the assessment of AIDS Risk in HIV-1-infected individuals
- PMID: 18776933
- PMCID: PMC2522281
- DOI: 10.1371/journal.pone.0003165
CCL3L1-CCR5 genotype improves the assessment of AIDS Risk in HIV-1-infected individuals
Abstract
Background: Whether vexing clinical decision-making dilemmas can be partly addressed by recent advances in genomics is unclear. For example, when to initiate highly active antiretroviral therapy (HAART) during HIV-1 infection remains a clinical dilemma. This decision relies heavily on assessing AIDS risk based on the CD4+ T cell count and plasma viral load. However, the trajectories of these two laboratory markers are influenced, in part, by polymorphisms in CCR5, the major HIV coreceptor, and the gene copy number of CCL3L1, a potent CCR5 ligand and HIV-suppressive chemokine. Therefore, we determined whether accounting for both genetic and laboratory markers provided an improved means of assessing AIDS risk.
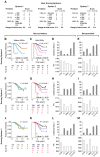
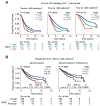
Methods and findings: In a prospective, single-site, ethnically-mixed cohort of 1,132 HIV-positive subjects, we determined the AIDS risk conveyed by the laboratory and genetic markers separately and in combination. Subjects were assigned to a low, moderate or high genetic risk group (GRG) based on variations in CCL3L1 and CCR5. The predictive value of the CCL3L1-CCR5 GRGs, as estimated by likelihood ratios, was equivalent to that of the laboratory markers. GRG status also predicted AIDS development when the laboratory markers conveyed a contrary risk. Additionally, in two separate and large groups of HIV+ subjects from a natural history cohort, the results from additive risk-scoring systems and classification and regression tree (CART) analysis revealed that the laboratory and CCL3L1-CCR5 genetic markers together provided more prognostic information than either marker alone. Furthermore, GRGs independently predicted the time interval from seroconversion to CD4+ cell count thresholds used to guide HAART initiation.
Conclusions: The combination of the laboratory and genetic markers captures a broader spectrum of AIDS risk than either marker alone. By tracking a unique aspect of AIDS risk distinct from that captured by the laboratory parameters, CCL3L1-CCR5 genotypes may have utility in HIV clinical management. These findings illustrate how genomic information might be applied to achieve practical benefits of personalized medicine.
Conflict of interest statement
Figures

References
-
- Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. Jama. 2006;296:815–826. - PubMed
-
- Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–3026. - PubMed
-
- Xin X, Shioda T, Kato A, Liu H, Sakai Y, et al. Enhanced anti-HIV-1 activity of CC-chemokine LD78beta, a non-allelic variant of MIP-1alpha/LD78alpha. FEBS Lett. 1999;457:219–222. - PubMed
-
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

