In GFP with high risk HPV-18E6 fusion protein expressed 293T and MCF-7 cells, the endogenous wild-type p53 could be transiently phosphorylated at multiple sites
- PMID: 18778462
- PMCID: PMC2546361
- DOI: 10.1186/1756-9966-27-35
In GFP with high risk HPV-18E6 fusion protein expressed 293T and MCF-7 cells, the endogenous wild-type p53 could be transiently phosphorylated at multiple sites
Abstract
Background: Infected cells recognize viral replication as a DNA damage stress and elicit the host surveillance mechanism to anti-virus infection. Modulation of the activity of tumor suppressor p53 is a key event in the replication of many viruses. They could manipulate p53 function through phosphorylation modification for their own purpose. But there is rarely research about p53 phosphorylation status in the context of HPV-E6. Therefore, we investigated whether p53 could be phosphorylated by HPV-E6.
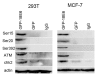
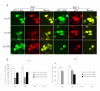
Methods: We used a mammalian green fluorescence protein (GFP) expression system to express HPV-18E6 with GFP fusion proteins (GFP-18E6) in wild-type (wt) p53 cell lines, such as 293T and MCF-7 cells to trace the traffic and subcellular location of E6 protein. By immunofluorescence technique and immunoblotting, we determined the positive phosphorylated sites of p53 and observed the distribution of phosphorylated p53 in the context of GFP-18E6.
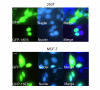
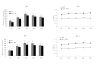
Results: GFP-18E6 was predominantly located in nuclei of wt p53 cell lines, and it could induce transient phosphorylation of p53 at multiple sites, such as Ser15, Ser20, and Ser392. All the three sites of phosphorylated p53s were localized in nuclei together with GFP-18E6.
Conclusion: In GFP with high risk HPV-18E6 fusion protein expressed 293T and MCF-7 cells, the endogenous wt p53 could be transiently phosphorylated at multiple sites.
Figures
Similar articles
-
HPV-16E6 can induce multiple site phosphorylation of p53.Oncol Rep. 2009 Feb;21(2):371-7. Oncol Rep. 2009. PMID: 19148510
-
The location of endogenous wild-type p53 protein in 293T and HEK293 cells expressing low-risk HPV-6E6 fusion protein with GFP.Acta Biochim Biophys Sin (Shanghai). 2010 Mar 15;42(3):230-5. doi: 10.1093/abbs/gmq009. Acta Biochim Biophys Sin (Shanghai). 2010. PMID: 20213049
-
GFP/HPV-16E6 fusion protein induces apoptosis in MCF-7 and 293T cells using a transient expression system.Oncol Rep. 2012 Nov;28(5):1673-80. doi: 10.3892/or.2012.1976. Epub 2012 Aug 22. Oncol Rep. 2012. PMID: 22922869
-
Cellular distribution of tumour suppressor protein p53 and high-risk human papillomavirus (HPV)-18 E6 fusion protein in wild-type p53 cell lines.J Int Med Res. 2008 Sep-Oct;36(5):1015-21. doi: 10.1177/147323000803600519. J Int Med Res. 2008. PMID: 18831896
-
Two different HPV-11E6 fusion proteins trap p53 in the cytoplasm and induce apoptosis.Cancer Biol Ther. 2008 Dec;7(12):1909-15. doi: 10.4161/cbt.7.12.6941. Cancer Biol Ther. 2008. PMID: 18981731
Cited by
-
Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis.J Exp Clin Cancer Res. 2010 Aug 11;29(1):110. doi: 10.1186/1756-9966-29-110. J Exp Clin Cancer Res. 2010. PMID: 20701792 Free PMC article. Review.
References
-
- Gumus M, Yumuk PF, Salepci T, Aliustaoglu M, Dane F, Ekenel M, Basaran G, Kaya H, Barisik N, Turhal NS. HPV DNA frequency and subset analysis in human breast cancer patients' normal and tumoral tissue samples. J Exp Clin Cancer Res. 2006;25:515–521. - PubMed
-
- Kehmeier E, Rühl H, Voland B, Stöppler MC. Cellular steady-state levels of "high risk" but not "low risk" human papillomavirus (HPV) E6 proteins are increased by inhibition of proteasome-dependent degradation independent of their p53- and E6AP-binding capabilities. Virology. 2002;299:72–87. doi: 10.1006/viro.2002.1502. - DOI - PubMed
-
- Indinnimeo M, Cicchini C, Stazi A, Giarnieri E, French D, Limiti MR, Ghini C, Vecchione A. Human papillomavirus infection and p53 nuclear overexpression in anal canal carcinoma. J Exp Clin Cancer Res. 1999;18:47–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

