DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0
- PMID: 18778471
- PMCID: PMC2542995
- DOI: 10.1186/1752-153X-2-18
DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0
Abstract
Background: Small-molecule docking is an important tool in studying receptor-ligand interactions and in identifying potential drug candidates. Previously, we developed a software tool (DOVIS) to perform large-scale virtual screening of small molecules in parallel on Linux clusters, using AutoDock 3.05 as the docking engine. DOVIS enables the seamless screening of millions of compounds on high-performance computing platforms. In this paper, we report significant advances in the software implementation of DOVIS 2.0, including enhanced screening capability, improved file system efficiency, and extended usability.
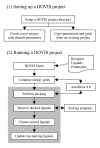
Implementation: To keep DOVIS up-to-date, we upgraded the software's docking engine to the more accurate AutoDock 4.0 code. We developed a new parallelization scheme to improve runtime efficiency and modified the AutoDock code to reduce excessive file operations during large-scale virtual screening jobs. We also implemented an algorithm to output docked ligands in an industry standard format, sd-file format, which can be easily interfaced with other modeling programs. Finally, we constructed a wrapper-script interface to enable automatic rescoring of docked ligands by arbitrarily selected third-party scoring programs.
Conclusion: The significance of the new DOVIS 2.0 software compared with the previous version lies in its improved performance and usability. The new version makes the computation highly efficient by automating load balancing, significantly reducing excessive file operations by more than 95%, providing outputs that conform to industry standard sd-file format, and providing a general wrapper-script interface for rescoring of docked ligands. The new DOVIS 2.0 package is freely available to the public under the GNU General Public License.
Figures

References
-
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. - DOI
-
- OpenBabel http://openbabel.sourceforge.net
LinkOut - more resources
Full Text Sources

