Structure and evolution of the C. elegans embryonic endomesoderm network
- PMID: 18778800
- PMCID: PMC2688470
- DOI: 10.1016/j.bbagrm.2008.07.013
Structure and evolution of the C. elegans embryonic endomesoderm network
Abstract
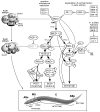
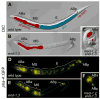
The specification of the Caenorhabditis elegans endomesoderm has been the subject of study for more than 15 years. Specification of the 4-cell stage endomesoderm precursor, EMS, occurs as a result of the activation of a transcription factor cascade that starts with SKN-1, coupled with input from the Wnt/beta-catenin asymmetry pathway through the nuclear effector POP-1. As development proceeds, transiently-expressed cell fate factors are succeeded by stable, tissue/organ-specific regulators. The pathway is complex and uses motifs found in all transcriptional networks. Here, the regulators that function in the C. elegans endomesoderm network are described. An examination of the motifs in the network suggests how they may have evolved from simpler gene interactions. Flexibility in the network is evident from the multitude of parallel functions that have been identified and from apparent changes in parts of the corresponding network in Caenorhabditis briggsae. Overall, the complexities of C. elegans endomesoderm specification build a picture of a network that is robust, complex, and still evolving.
Figures
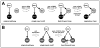
Similar articles
-
Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network.Dev Biol. 2010 Apr 15;340(2):209-21. doi: 10.1016/j.ydbio.2009.09.042. Epub 2009 Oct 7. Dev Biol. 2010. PMID: 19818340 Free PMC article.
-
Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans.Dev Biol. 2009 Jan 1;325(1):296-306. doi: 10.1016/j.ydbio.2008.10.001. Epub 2008 Oct 19. Dev Biol. 2009. PMID: 18977344 Free PMC article.
-
Endomesoderm specification in Caenorhabditis elegans and other nematodes.Bioessays. 2006 Oct;28(10):1010-22. doi: 10.1002/bies.20480. Bioessays. 2006. PMID: 16998834 Review.
-
The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development.Dev Biol. 2005 Sep 15;285(2):510-23. doi: 10.1016/j.ydbio.2005.06.022. Dev Biol. 2005. PMID: 16084508
-
The Caenorhabditis elegans intestine.Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review.
Cited by
-
Asymmetric Wnt Pathway Signaling Facilitates Stem Cell-Like Divisions via the Nonreceptor Tyrosine Kinase FRK-1 in Caenorhabditis elegans.Genetics. 2015 Nov;201(3):1047-60. doi: 10.1534/genetics.115.181412. Epub 2015 Sep 9. Genetics. 2015. PMID: 26358719 Free PMC article.
-
ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult.Dev Biol. 2009 Mar 15;327(2):551-65. doi: 10.1016/j.ydbio.2008.11.034. Epub 2008 Dec 9. Dev Biol. 2009. PMID: 19111532 Free PMC article.
-
The molecular basis of organ formation: insights from the C. elegans foregut.Annu Rev Cell Dev Biol. 2009;25:597-628. doi: 10.1146/annurev.cellbio.24.110707.175411. Annu Rev Cell Dev Biol. 2009. PMID: 19575642 Free PMC article. Review.
-
TORC2 signaling antagonizes SKN-1 to induce C. elegans mesendodermal embryonic development.Dev Biol. 2013 Dec 15;384(2):214-27. doi: 10.1016/j.ydbio.2013.08.011. Epub 2013 Aug 20. Dev Biol. 2013. PMID: 23973804 Free PMC article.
-
The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development.Development. 2009 Aug;136(16):2735-46. doi: 10.1242/dev.038307. Epub 2009 Jul 15. Development. 2009. PMID: 19605496 Free PMC article.
References
-
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. - PubMed
-
- Maduro MF. Endomesoderm specification in Caenorhabditis elegans and other nematodes. Bioessays. 2006;28:1010–1022. - PubMed
-
- Mizumoto K, Sawa H. Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell. 2007;12:287–299. - PubMed
-
- Schierenberg E. Three sons of fortune: early embryogenesis, evolution and ecology of nematodes. Bioessays. 2001;23:841–847. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

