Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes
- PMID: 18779312
- PMCID: PMC2718693
- DOI: 10.1124/dmd.108.023770
Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes
Abstract
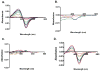
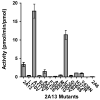
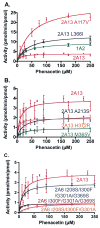
Cytochrome P450s (P450s) metabolize a large number of diverse substrates with specific regio- and stereospecificity. A number of compounds, including nicotine, cotinine, and aflatoxin B(1), are metabolites of the 94% identical CYP2A13 and CYP2A6 enzymes but at different rates. Phenacetin and 4-aminobiphenyl were identified as substrates of human cytochromes P450 1A2 and 2A13 but not of CYP2A6. The purpose of this study was to identify active site amino acids that are responsible for CYP2A substrate specificity using phenacetin as a structural probe. Ten amino acid residues that differ in the CYP2A13 and CYP2A6 active sites were exchanged between the two enzymes. Phenacetin binding revealed that the six substitution, CYP2A13 S208I, A213S, F300I, A301G, M365V, and G369S decreased phenacetin affinity. Although incorporation of individual CYP2A13 residues into CYP2A6 had little effect on this enzyme's very low levels of phenacetin metabolism, the combination of double, triple, and quadruple substitutions at positions 208, 300, 301, and 369 increasingly endowed CYP2A6 with the ability to metabolize phenacetin. Enzyme kinetics revealed that the CYP2A6 I208S/I300F/G301A/S369G mutant protein O-deethylated phenacetin with a K(m) of 10.3 muM and a k(cat) of 2.9 min(-1), which compare very favorably with those of CYP2A13 (K(m) of 10.7 muM and k(cat) of 3.8 min(-1)). A 2.15 A crystal structure of the mutant CYP2A6 I208S/I300F/G301A/S369G protein with phenacetin in the active site provided a structural rationale for the differences in phenacetin metabolism between CYP2A6 and CYP2A13.
Figures



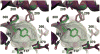
References
-
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. - PubMed
-
- Bao Z, He XY, Ding X, Prabhu S, Hong JY. Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab Dispos. 2005;33:258–261. - PubMed
-
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002.
-
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. - PubMed
-
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

