Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling
- PMID: 18779369
- PMCID: PMC2528591
- DOI: 10.1083/jcb.200802084
Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling
Abstract
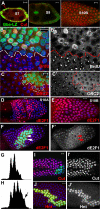
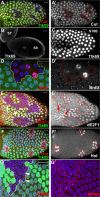
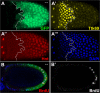
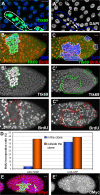
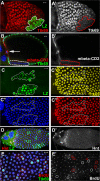
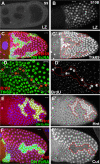
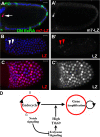
The developmental signals that regulate the switch from genome-wide DNA replication to site-specific amplification remain largely unknown. Drosophila melanogaster epithelial follicle cells, which begin synchronized chorion gene amplification after three rounds of endocycle, provide an excellent model for study of the endocycle/gene amplification (E/A) switch. Here, we report that down-regulation of Notch signaling and activation of ecdysone receptor (EcR) are required for the E/A switch in these cells. Extended Notch activity suppresses EcR activation and prevents exit from the endocycle. Tramtrack (Ttk), a zinc-finger protein essential for the switch, is regulated negatively by Notch and positively by EcR. Ttk overexpression stops endoreplication prematurely and alleviates the endocycle exit defect caused by extended Notch activity or removal of EcR function. Our results reveal a developmental pathway that includes down-regulation of Notch, activation of the EcR, up-regulation of Ttk to execute the E/A switch, and, for the first time, the genetic interaction between Notch and ecdysone signaling in regulation of cell cycle programs and differentiation.
Figures
Similar articles
-
Nuclear receptors linking physiology and germline stem cells in Drosophila.Vitam Horm. 2021;116:327-362. doi: 10.1016/bs.vh.2020.12.008. Vitam Horm. 2021. PMID: 33752824 Free PMC article. Review.
-
The Ecdysone and Notch Pathways Synergistically Regulate Cut at the Dorsal-Ventral Boundary in Drosophila Wing Discs.J Genet Genomics. 2016 Apr 20;43(4):179-86. doi: 10.1016/j.jgg.2016.03.002. Epub 2016 Mar 17. J Genet Genomics. 2016. PMID: 27117286 Free PMC article.
-
Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation.Dev Cell. 2007 Mar;12(3):431-42. doi: 10.1016/j.devcel.2007.02.003. Dev Cell. 2007. PMID: 17336908 Free PMC article.
-
Regulation of broad by the Notch pathway affects timing of follicle cell development.Dev Biol. 2014 Aug 1;392(1):52-61. doi: 10.1016/j.ydbio.2014.04.024. Epub 2014 May 9. Dev Biol. 2014. PMID: 24815210 Free PMC article.
-
Ecdysone signaling in adult Drosophila melanogaster.J Insect Physiol. 2012 Mar;58(3):293-302. doi: 10.1016/j.jinsphys.2012.01.013. Epub 2012 Jan 28. J Insect Physiol. 2012. PMID: 22310011 Review.
Cited by
-
Nuclear receptors linking physiology and germline stem cells in Drosophila.Vitam Horm. 2021;116:327-362. doi: 10.1016/bs.vh.2020.12.008. Vitam Horm. 2021. PMID: 33752824 Free PMC article. Review.
-
Regulation of pattern formation and gene amplification during Drosophila oogenesis by the miR-318 microRNA.Genetics. 2015 May;200(1):255-65. doi: 10.1534/genetics.115.174748. Epub 2015 Mar 17. Genetics. 2015. PMID: 25786856 Free PMC article.
-
CoREST acts as a positive regulator of Notch signaling in the follicle cells of Drosophila melanogaster.J Cell Sci. 2012 Jan 15;125(Pt 2):399-410. doi: 10.1242/jcs.089797. Epub 2012 Feb 13. J Cell Sci. 2012. PMID: 22331351 Free PMC article.
-
Tramtrack acts during late pupal development to direct ant caste identity.PLoS Genet. 2021 Sep 22;17(9):e1009801. doi: 10.1371/journal.pgen.1009801. eCollection 2021 Sep. PLoS Genet. 2021. PMID: 34550980 Free PMC article.
-
DNA Replication Control During Drosophila Development: Insights into the Onset of S Phase, Replication Initiation, and Fork Progression.Genetics. 2017 Sep;207(1):29-47. doi: 10.1534/genetics.115.186627. Genetics. 2017. PMID: 28874453 Free PMC article. Review.
References
-
- Aggarwal, B.D., and B.R. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 430:372–376. - PubMed
-
- Assa-Kunik, E., I.L. Torres, E.D. Schejter, D.S. Johnston, and B.Z. Shilo. 2007. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development. 134:1161–1169. - PubMed
-
- Audibert, A., F. Simon, and M. Gho. 2005. Cell cycle diversity involves differential regulation of Cyclin E activity in the Drosophila bristle cell lineage. Development. 132:2287–2297. - PubMed
-
- Badenhorst, P. 2001. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development. 128:4093–4101. - PubMed
-
- Beall, E.L., J.R. Manak, S. Zhou, M. Bell, J.S. Lipsick, and M.R. Botchan. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 420:833–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

