Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation
- PMID: 18779370
- PMCID: PMC2528577
- DOI: 10.1083/jcb.200805140
Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation
Abstract
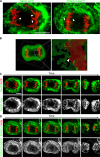
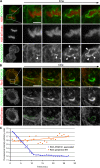
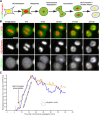
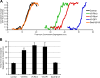
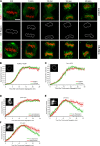
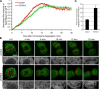
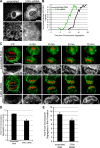
During mitosis in metazoans, segregated chromosomes become enclosed by the nuclear envelope (NE), a double membrane that is continuous with the endoplasmic reticulum (ER). Recent in vitro data suggest that NE formation occurs by chromatin-mediated reorganization of the tubular ER; however, the basic principles of such a membrane-reshaping process remain uncharacterized. Here, we present a quantitative analysis of nuclear membrane assembly in mammalian cells using time-lapse microscopy. From the initial recruitment of ER tubules to chromatin, the formation of a membrane-enclosed, transport-competent nucleus occurs within approximately 12 min. Overexpression of the ER tubule-forming proteins reticulon 3, reticulon 4, and DP1 inhibits NE formation and nuclear expansion, whereas their knockdown accelerates nuclear assembly. This suggests that the transition from membrane tubules to sheets is rate-limiting for nuclear assembly. Our results provide evidence that ER-shaping proteins are directly involved in the reconstruction of the nuclear compartment and that morphological restructuring of the ER is the principal mechanism of NE formation in vivo.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

