Accelerated evolution of resistance in multidrug environments
- PMID: 18779569
- PMCID: PMC2544564
- DOI: 10.1073/pnas.0805965105
Accelerated evolution of resistance in multidrug environments
Abstract
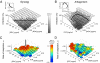
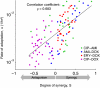
The emergence of resistance during multidrug chemotherapy impedes the treatment of many human diseases, including malaria, TB, HIV, and cancer. Although certain combination therapies have long been known to be more effective in curing patients than single drugs, the impact of such treatments on the evolution of drug resistance is unclear. In particular, very little is known about how the evolution of resistance is affected by the nature of the interactions--synergy or antagonism--between drugs. Here we directly measure the effect of various inhibitory and subinhibitory drug combinations on the rate of adaptation. We develop an automated assay for monitoring the parallel evolution of hundreds of Escherichia coli populations in a two-dimensional grid of drug gradients over many generations. We find a correlation between synergy and the rate of adaptation, whereby evolution in more synergistic drug combinations, typically preferred in clinical settings, is faster than evolution in antagonistic combinations. We also find that resistance to some synergistic combinations evolves faster than resistance to individual drugs. The accelerated evolution may be due to a larger selective advantage for resistance mutations in synergistic treatments. We describe a simple geometric model in which mutations conferring resistance to one drug of a synergistic pair prevent not only the inhibitory effect of that drug but also its enhancing effect on the other drug. Future study of the profound impact that synergy and other drug-pair properties can have on the rate of adaptation may suggest new treatment strategies for combating the spread of antibiotic resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. - PubMed
-
- Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: American Society for Microbiology Press; 2003.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

