In pursuit of virtual lead optimization: pruning ensembles of receptor structures for increased efficiency and accuracy during docking
- PMID: 18781587
- PMCID: PMC2649978
- DOI: 10.1002/prot.22214
In pursuit of virtual lead optimization: pruning ensembles of receptor structures for increased efficiency and accuracy during docking
Abstract
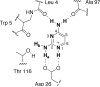
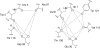
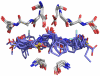
Representing receptors as ensembles of protein conformations during docking is a powerful method to approximate protein flexibility and increase the accuracy of the resulting ranked list of compounds. Unfortunately, docking compounds against a large number of ensemble members can increase computational cost and time investment. In this article, we present an efficient method to evaluate and select the most contributive ensemble members prior to docking for targets with a conserved core of residues that bind a ligand moiety. We observed that ensemble members that preserve the geometry of the active site core are most likely to place ligands in the active site with a conserved orientation, generally rank ligands correctly and increase interactions with the receptor. A relative distance approach is used to quantify the preservation of the three-dimensional interatomic distances of the conserved ligand-binding atoms and prune large ensembles quickly. In this study, we investigate dihydrofolate reductase as an example of a protein with a conserved core; however, this method for accurately selecting relevant ensemble members a priori can be applied to any system with a conserved ligand-binding core, including HIV-1 protease, kinases, and acetylcholinesterase. Representing a drug target as a pruned ensemble during in silico screening should increase the accuracy and efficiency of high-throughput analyses of lead analogs.
(c) 2008 Wiley-Liss, Inc.
Figures


Similar articles
-
Scoring ensembles of docked protein:ligand interactions for virtual lead optimization.J Chem Inf Model. 2009 Dec;49(12):2813-9. doi: 10.1021/ci9003078. J Chem Inf Model. 2009. PMID: 19950979 Free PMC article.
-
In pursuit of virtual lead optimization: the role of the receptor structure and ensembles in accurate docking.Proteins. 2008 Nov 15;73(3):566-80. doi: 10.1002/prot.22081. Proteins. 2008. PMID: 18473360 Free PMC article.
-
Towards in silico lead optimization: scores from ensembles of protein/ligand conformations reliably correlate with biological activity.Proteins. 2007 Feb 1;66(2):375-87. doi: 10.1002/prot.21201. Proteins. 2007. PMID: 17078091
-
Flexible receptor docking for drug discovery.Expert Opin Drug Discov. 2015;10(11):1189-200. doi: 10.1517/17460441.2015.1078308. Epub 2015 Aug 26. Expert Opin Drug Discov. 2015. PMID: 26313123 Review.
-
Challenges of target/compound data integration from disease to chemistry: a case study of dihydrofolate reductase inhibitors.Curr Drug Discov Technol. 2005 Jun;2(2):75-87. doi: 10.2174/1570163054064675. Curr Drug Discov Technol. 2005. PMID: 16472232 Review.
Cited by
-
How to choose relevant multiple receptor conformations for virtual screening: a test case of Cdk2 and normal mode analysis.Eur Biophys J. 2010 Aug;39(9):1365-72. doi: 10.1007/s00249-010-0592-0. Epub 2010 Mar 18. Eur Biophys J. 2010. PMID: 20237920
-
A comparative study of AutoDock and PMF scoring performances, and SAR of 2-substituted pyrazolotriazolopyrimidines and 4-substituted pyrazolopyrimidines as potent xanthine oxidase inhibitors.J Comput Aided Mol Des. 2010 Jan;24(1):57-75. doi: 10.1007/s10822-009-9314-z. Epub 2009 Dec 29. J Comput Aided Mol Des. 2010. PMID: 20039101
-
A Multi-scale Computational Platform to Mechanistically Assess the Effect of Genetic Variation on Drug Responses in Human Erythrocyte Metabolism.PLoS Comput Biol. 2016 Jul 28;12(7):e1005039. doi: 10.1371/journal.pcbi.1005039. eCollection 2016 Jul. PLoS Comput Biol. 2016. PMID: 27467583 Free PMC article.
-
Scoring ensembles of docked protein:ligand interactions for virtual lead optimization.J Chem Inf Model. 2009 Dec;49(12):2813-9. doi: 10.1021/ci9003078. J Chem Inf Model. 2009. PMID: 19950979 Free PMC article.
-
Efficient incorporation of protein flexibility and dynamics into molecular docking simulations.Biochemistry. 2011 Jul 19;50(28):6157-69. doi: 10.1021/bi2004558. Epub 2011 Jun 22. Biochemistry. 2011. PMID: 21678954 Free PMC article. Review.
References
-
- Carlson HA. Protein flexibility and drug design: how to hit a moving target. Curr Opin Chem Biol. 2002;6(4):447–452. - PubMed
-
- Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000;43(11):2100–2114. - PubMed
-
- Popov VM, Yee WA, Anderson AC. Towards in silico lead optimization: scores from ensembles of protein/ligand conformations reliably correlate with biological activity. Proteins. 2007;66(2):375–387. - PubMed
-
- Rao S, Sanschagrin PC, Greenwood JR, Repasky MP, Sherman W, Farid R. Improving database enrichment through ensemble docking. J Comput Aided Mol Des. 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

