Metabolic innovations towards the human lineage
- PMID: 18782449
- PMCID: PMC2553087
- DOI: 10.1186/1471-2148-8-247
Metabolic innovations towards the human lineage
Abstract
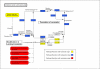
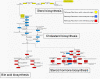
Background: We describe a function-driven approach to the analysis of metabolism which takes into account the phylogenetic origin of biochemical reactions to reveal subtle lineage-specific metabolic innovations, undetectable by more traditional methods based on sequence comparison. The origins of reactions and thus entire pathways are inferred using a simple taxonomic classification scheme that describes the evolutionary course of events towards the lineage of interest. We investigate the evolutionary history of the human metabolic network extracted from a metabolic database, construct a network of interconnected pathways and classify this network according to the taxonomic categories representing eukaryotes, metazoa and vertebrates.
Results: It is demonstrated that lineage-specific innovations correspond to reactions and pathways associated with key phenotypic changes during evolution, such as the emergence of cellular organelles in eukaryotes, cell adhesion cascades in metazoa and the biosynthesis of complex cell-specific biomolecules in vertebrates.
Conclusion: This phylogenetic view of metabolic networks puts gene innovations within an evolutionary context, demonstrating how the emergence of a phenotype in a lineage provides a platform for the development of specialized traits.
Figures


References
-
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S. et al.The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415(6874):871–880. doi: 10.1038/nature724. - DOI - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al.Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

