The pathway via D-galacturonate/L-galactonate is significant for ascorbate biosynthesis in Euglena gracilis: identification and functional characterization of aldonolactonase
- PMID: 18782759
- PMCID: PMC2662179
- DOI: 10.1074/jbc.M803930200
The pathway via D-galacturonate/L-galactonate is significant for ascorbate biosynthesis in Euglena gracilis: identification and functional characterization of aldonolactonase
Abstract
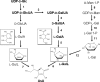
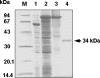
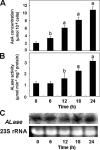
We have previously proposed that Euglena gracilis possesses a pathway for the production of ascorbate (AsA) through d-galacturonate/L-galactonate as representative intermediates ( Shigeoka, S., Nakano, Y., and Kitaoka, S. (1979) J. Nutr. Sci. Vitaminol. 25, 299-307 ). However, genetic evidence proving that the pathway exists has not been obtained yet. We report here the identification of a gene encoding aldonolactonase, which catalyzes a penultimate step of the biosynthesis of AsA in Euglena. By a BLAST search, we identified one candidate for the enzyme having significant sequence identity with rat gluconolactonase, a key enzyme for the production of AsA via d-glucuronate in animals. The purified recombinant aldonolactonase expressed in Escherichia coli catalyzed the reversible reaction of L-galactonate and L-galactono-1,4-lactone with zinc ion as a cofactor. The apparent K(m) values for L-galactonate and L-galactono-1,4-lactone were 1.55 +/- 0.3 and 1.67 +/- 0.39 mm, respectively. The cell growth of Euglena was arrested by silencing the expression of aldonolactonase through RNA interference and then restored to the normal state by supplementation with L-galactono-1,4-lactone. Euglena cells accumulated more AsA on supplementation with d-galacturonate than d-glucuronate. The present results indicate that aldonolactonase is significant for the biosynthesis of AsA in Euglena cells, which predominantly utilize the pathwayviad-galacturonate/L-galactonate. The identification of aldonolactonase provides the first insight into the biosynthesis of AsA via uronic acids as the intermediate in photosynthetic algae including Euglena.
Figures





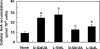
Similar articles
-
Metabolic engineering of the fungal D-galacturonate pathway for L-ascorbic acid production.Microb Cell Fact. 2015 Jan 8;14:2. doi: 10.1186/s12934-014-0184-2. Microb Cell Fact. 2015. PMID: 25566698 Free PMC article.
-
Functional characterization of D-galacturonic acid reductase, a key enzyme of the ascorbate biosynthesis pathway, from Euglena gracilis.Biosci Biotechnol Biochem. 2006 Nov;70(11):2720-6. doi: 10.1271/bbb.60327. Epub 2006 Nov 7. Biosci Biotechnol Biochem. 2006. PMID: 17090924
-
The d-mannose/l-galactose pathway is the dominant ascorbate biosynthetic route in the moss Physcomitrium patens.Plant J. 2021 Sep;107(6):1724-1738. doi: 10.1111/tpj.15413. Epub 2021 Aug 10. Plant J. 2021. PMID: 34245628
-
L-ascorbic acid biosynthesis.Vitam Horm. 2001;61:241-66. doi: 10.1016/s0083-6729(01)61008-2. Vitam Horm. 2001. PMID: 11153268 Review.
-
Recent progress on the characterization of aldonolactone oxidoreductases.Plant Physiol Biochem. 2016 Jan;98:171-85. doi: 10.1016/j.plaphy.2015.11.017. Epub 2015 Nov 27. Plant Physiol Biochem. 2016. PMID: 26696130 Free PMC article. Review.
Cited by
-
Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C.Plant Physiol. 2009 Jun;150(2):596-605. doi: 10.1104/pp.109.136929. Epub 2009 Apr 15. Plant Physiol. 2009. PMID: 19369590 Free PMC article.
-
Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes.PLoS Genet. 2018 Jul 30;14(7):e1007533. doi: 10.1371/journal.pgen.1007533. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30059538 Free PMC article.
-
Transgene Expression in Microalgae-From Tools to Applications.Front Plant Sci. 2016 Apr 22;7:505. doi: 10.3389/fpls.2016.00505. eCollection 2016. Front Plant Sci. 2016. PMID: 27148328 Free PMC article. Review.
-
Genomics Reveals the Metabolic Potential and Functions in the Redistribution of Dissolved Organic Matter in Marine Environments of the Genus Thalassotalea.Microorganisms. 2020 Sep 14;8(9):1412. doi: 10.3390/microorganisms8091412. Microorganisms. 2020. PMID: 32937826 Free PMC article.
-
Diverse Biosynthetic Pathways and Protective Functions against Environmental Stress of Antioxidants in Microalgae.Plants (Basel). 2021 Jun 19;10(6):1250. doi: 10.3390/plants10061250. Plants (Basel). 2021. PMID: 34205386 Free PMC article. Review.
References
-
- Shigeoka, S., Nakano, Y., and Kitaoka, S. (1979) J. Nutr. Sci. Vitaminol. 25 299-307 - PubMed
-
- Davey, M. W., Van Montagu, M., Inze, D., Sanmartin, M., Kanellis, A., Smirnoff, N., Benzie, I. J. J., Strain, J. J., Favell, D., and Fletcher, J. (2000) J. Sci. Food Agric. 80 825-860
-
- Smirnoff, N. (2000) Curr. Opin. Plant Biol. 3229 -235 - PubMed
-
- Ishikawa, T., Dowdle, J., and Smirnoff, N. (2006) Physiol. Plant. 126343 -355
-
- Nishikimi, M., and Yagi, K. (1996) Subcell. Biochem. 2517 -39 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

