Targeting of hFis1 to peroxisomes is mediated by Pex19p
- PMID: 18782765
- PMCID: PMC2662177
- DOI: 10.1074/jbc.M803332200
Targeting of hFis1 to peroxisomes is mediated by Pex19p
Abstract
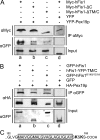
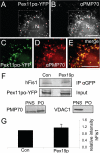
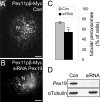
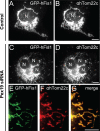
The processes of peroxisome formation and proliferation are still a matter of debate. We have previously shown that peroxisomes share some components of their division machinery with mitochondria. hFis1, a tail-anchored membrane protein, regulates the membrane fission of both organelles by DLP1/Drp1 recruitment, but nothing is known about the mechanisms of the dual targeting of hFis1. Here we demonstrate for the first time that peroxisomal targeting of hFis1 depends on Pex19p, a peroxisomal membrane protein import factor. hFis1/Pex19p binding was demonstrated by expression and co-immunoprecipitation studies. Using mutated versions of hFis1 an essential binding region for Pex19p was located within the last 26 C-terminal amino acids of hFis1, which are required for proper targeting to both mitochondria and peroxisomes. The basic amino acids in the very C terminus are not essential for Pex19p binding and peroxisomal targeting, but are instead required for mitochondrial targeting. Silencing of Pex19p by small interference RNA reduced the targeting of hFis1 to peroxisomes, but not to mitochondria. In contrast, overexpression of Pex19p alone was not sufficient to shift the targeting of hFis1 to peroxisomes. Our findings indicate that targeting of hFis1 to peroxisomes and mitochondria are independent events and support a direct, Pex19p-dependent targeting of peroxisomal tail-anchored proteins.
Figures




Similar articles
-
A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells.Mol Biol Cell. 2005 Nov;16(11):5077-86. doi: 10.1091/mbc.e05-02-0159. Epub 2005 Aug 17. Mol Biol Cell. 2005. PMID: 16107562 Free PMC article.
-
Assembly of Peroxisomal Membrane Proteins via the Direct Pex19p-Pex3p Pathway.Traffic. 2016 Apr;17(4):433-55. doi: 10.1111/tra.12376. Epub 2016 Feb 23. Traffic. 2016. PMID: 26777132
-
The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step.J Biol Chem. 2006 Nov 10;281(45):34492-502. doi: 10.1074/jbc.M607183200. Epub 2006 Sep 15. J Biol Chem. 2006. PMID: 16980692
-
Shared components of mitochondrial and peroxisomal division.Biochim Biophys Acta. 2006 May-Jun;1763(5-6):531-41. doi: 10.1016/j.bbamcr.2006.01.004. Epub 2006 Feb 2. Biochim Biophys Acta. 2006. PMID: 16487606 Review.
-
Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions.Biochim Biophys Acta. 2006 Dec;1763(12):1639-46. doi: 10.1016/j.bbamcr.2006.09.030. Epub 2006 Sep 26. Biochim Biophys Acta. 2006. PMID: 17069900 Review.
Cited by
-
Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway.J Cell Biol. 2013 Mar 4;200(5):651-66. doi: 10.1083/jcb.201211077. J Cell Biol. 2013. PMID: 23460677 Free PMC article.
-
The Ways of Tails: the GET Pathway and more.Protein J. 2019 Jun;38(3):289-305. doi: 10.1007/s10930-019-09845-4. Protein J. 2019. PMID: 31203484 Review.
-
Mitochondrial Fission Protein 1: Emerging Roles in Organellar Form and Function in Health and Disease.Front Endocrinol (Lausanne). 2021 Mar 25;12:660095. doi: 10.3389/fendo.2021.660095. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33841340 Free PMC article. Review.
-
A bacteria-derived tail anchor localizes to peroxisomes in yeast and mammalian cells.Sci Rep. 2018 Nov 6;8(1):16374. doi: 10.1038/s41598-018-34646-7. Sci Rep. 2018. PMID: 30401812 Free PMC article.
-
The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria.Plant Cell. 2011 Dec;23(12):4446-61. doi: 10.1105/tpc.111.090142. Epub 2011 Dec 6. Plant Cell. 2011. PMID: 22147290 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

