Triggering protein folding within the GroEL-GroES complex
- PMID: 18782766
- PMCID: PMC2581556
- DOI: 10.1074/jbc.M802898200
Triggering protein folding within the GroEL-GroES complex
Abstract
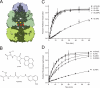
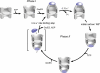
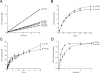
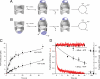
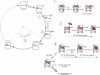
The folding of many proteins depends on the assistance of chaperonins like GroEL and GroES and involves the enclosure of substrate proteins inside an internal cavity that is formed when GroES binds to GroEL in the presence of ATP. Precisely how assembly of the GroEL-GroES complex leads to substrate protein encapsulation and folding remains poorly understood. Here we use a chemically modified mutant of GroEL (EL43Py) to uncouple substrate protein encapsulation from release and folding. Although EL43Py correctly initiates a substrate protein encapsulation reaction, this mutant stalls in an intermediate allosteric state of the GroEL ring, which is essential for both GroES binding and the forced unfolding of the substrate protein. This intermediate conformation of the GroEL ring possesses simultaneously high affinity for both GroES and non-native substrate protein, thus preventing escape of the substrate protein while GroES binding and substrate protein compaction takes place. Strikingly, assembly of the folding-active GroEL-GroES complex appears to involve a strategic delay in ATP hydrolysis that is coupled to disassembly of the old, ADP-bound GroEL-GroES complex on the opposite ring.
Figures


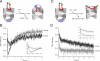
Similar articles
-
Asymmetrical interaction of GroEL and GroES in the ATPase cycle of assisted protein folding.Science. 1995 Aug 11;269(5225):836-41. doi: 10.1126/science.7638601. Science. 1995. PMID: 7638601
-
Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction.Cell. 1996 Feb 9;84(3):481-90. doi: 10.1016/s0092-8674(00)81293-3. Cell. 1996. PMID: 8608602
-
Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant.J Biol Chem. 2008 Aug 29;283(35):23774-81. doi: 10.1074/jbc.M802542200. Epub 2008 Jun 20. J Biol Chem. 2008. PMID: 18567584 Free PMC article.
-
GroEL and the GroEL-GroES Complex.Subcell Biochem. 2017;83:483-504. doi: 10.1007/978-3-319-46503-6_17. Subcell Biochem. 2017. PMID: 28271487 Review.
-
Reaction Cycle of Chaperonin GroEL via Symmetric "Football" Intermediate.J Mol Biol. 2015 Sep 11;427(18):2912-8. doi: 10.1016/j.jmb.2015.04.007. Epub 2015 Apr 18. J Mol Biol. 2015. PMID: 25900372 Review.
Cited by
-
How do chaperonins fold protein?Biophysics (Nagoya-shi). 2015 Apr 1;11:93-102. doi: 10.2142/biophysics.11.93. eCollection 2015. Biophysics (Nagoya-shi). 2015. PMID: 27493521 Free PMC article. Review.
-
GroEL Mediates Folding of Bacillus anthracis Serine/Threonine Protein Kinase, PrkC.Indian J Microbiol. 2018 Dec;58(4):520-524. doi: 10.1007/s12088-018-0744-y. Epub 2018 May 30. Indian J Microbiol. 2018. PMID: 30262963 Free PMC article.
-
Polypeptide in the chaperonin cage partly protrudes out and then folds inside or escapes outside.EMBO J. 2010 Dec 1;29(23):4008-19. doi: 10.1038/emboj.2010.262. Epub 2010 Oct 19. EMBO J. 2010. PMID: 20959808 Free PMC article.
-
Structure of GroEL in complex with an early folding intermediate of alanine glyoxylate aminotransferase.J Biol Chem. 2010 Feb 26;285(9):6371-6. doi: 10.1074/jbc.M109.062471. Epub 2010 Jan 7. J Biol Chem. 2010. PMID: 20056599 Free PMC article.
-
How soluble misfolded proteins bypass chaperones at the molecular level.Nat Commun. 2023 Jun 21;14(1):3689. doi: 10.1038/s41467-023-38962-z. Nat Commun. 2023. PMID: 37344452 Free PMC article.
References
-
- Young, J. C., Agashe, V. R., Siegers, K., and Hartl, F. U. (2004) Nat. Rev. Mol. Cell. Biol. 5 781-791 - PubMed
-
- Fenton, W. A., and Horwich, A. L. (2003) Q. Rev. Biophys. 36 229-256 - PubMed
-
- Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D. C., Joachimiak, A., Horwich, A. L., and Sigler, P. B. (1994) Nature 371 578-586 - PubMed
-
- Rye, H. S., Burston, S. G., Fenton, W. A., Beechem, J. M., Xu, Z., Sigler, P. B., and Horwich, A. L. (1997) Nature 388 792-798 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

