The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity
- PMID: 18782831
- PMCID: PMC2566870
- DOI: 10.1093/nar/gkn581
The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity
Abstract
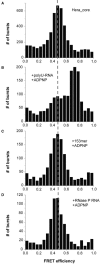
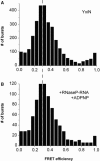
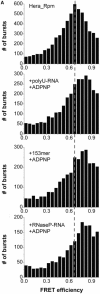
DEAD box helicases use the energy of ATP hydrolysis to remodel RNA structures or RNA/protein complexes. They share a common helicase core with conserved signature motifs, and additional domains may confer substrate specificity. Identification of a specific substrate is crucial towards understanding the physiological role of a helicase. RNA binding and ATPase stimulation are necessary, but not sufficient criteria for a bona fide helicase substrate. Here, we report single molecule FRET experiments that identify fragments of the 23S rRNA comprising hairpin 92 and RNase P RNA as substrates for the Thermus thermophilus DEAD box helicase Hera. Both substrates induce a switch to the closed conformation of the helicase core and stimulate the intrinsic ATPase activity of Hera. Binding of these RNAs is mediated by the Hera C-terminal domain, but does not require a previously proposed putative RNase P motif within this domain. ATP-dependent unwinding of a short helix adjacent to hairpin 92 in the ribosomal RNA suggests a specific role for Hera in ribosome assembly, analogously to the Escherichia coli and Bacillus subtilis helicases DbpA and YxiN. In addition, the specificity of Hera for RNase P RNA may be required for RNase P RNA folding or RNase P assembly.
Figures



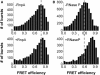

References
-
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. - PubMed
-
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429.
-
- Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. - PubMed
-
- Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

