LOMA: a fast method to generate efficient tagged-random primers despite amplification bias of random PCR on pathogens
- PMID: 18783594
- PMCID: PMC2553803
- DOI: 10.1186/1471-2105-9-368
LOMA: a fast method to generate efficient tagged-random primers despite amplification bias of random PCR on pathogens
Abstract
Background: Pathogen detection using DNA microarrays has the potential to become a fast and comprehensive diagnostics tool. However, since pathogen detection chips currently utilize random primers rather than specific primers for the RT-PCR step, bias inherent in random PCR amplification becomes a serious problem that causes large inaccuracies in hybridization signals.
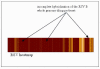
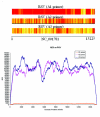
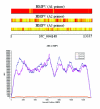
Results: In this paper, we study how the efficiency of random PCR amplification affects hybridization signals. We describe a model that predicts the amplification efficiency of a given random primer on a target viral genome. The prediction allows us to filter false-negative probes of the genome that lie in regions of poor random PCR amplification and improves the accuracy of pathogen detection. Subsequently, we propose LOMA, an algorithm to generate random primers that have good amplification efficiency. Wet-lab validation showed that the generated random primers improve the amplification efficiency significantly.
Conclusion: The blind use of a random primer with attached universal tag (random-tagged primer) in a PCR reaction on a pathogen sample may not lead to a successful amplification. Thus, the design of random-tagged primers is an important consideration when performing PCR.
Figures



Similar articles
-
Dual-genome primer design for construction of DNA microarrays.Bioinformatics. 2005 Feb 1;21(3):325-32. doi: 10.1093/bioinformatics/bti001. Epub 2004 Aug 27. Bioinformatics. 2005. PMID: 15333463
-
Novel computational methods for increasing PCR primer design effectiveness in directed sequencing.BMC Bioinformatics. 2008 Apr 11;9:191. doi: 10.1186/1471-2105-9-191. BMC Bioinformatics. 2008. PMID: 18405373 Free PMC article.
-
Microarray synthesis through multiple-use PCR primer design.Bioinformatics. 2002;18 Suppl 1:S128-35. doi: 10.1093/bioinformatics/18.suppl_1.s128. Bioinformatics. 2002. PMID: 12169540
-
MultiPrimer: a system for microarray PCR primer design.Methods Mol Biol. 2007;402:305-14. doi: 10.1007/978-1-59745-528-2_15. Methods Mol Biol. 2007. PMID: 17951802 Review.
-
Applications of random PCR.Cell Mol Biol (Noisy-le-grand). 1995 Jul;41(5):653-70. Cell Mol Biol (Noisy-le-grand). 1995. PMID: 7580845 Review.
Cited by
-
Recent advances and application of whole genome amplification in molecular diagnosis and medicine.MedComm (2020). 2022 Feb 3;3(1):e116. doi: 10.1002/mco2.116. eCollection 2022 Mar. MedComm (2020). 2022. PMID: 35281794 Free PMC article. Review.
-
Expecting the unexpected: nucleic acid-based diagnosis and discovery of emerging viruses.Expert Rev Mol Diagn. 2011 May;11(4):409-23. doi: 10.1586/erm.11.24. Expert Rev Mol Diagn. 2011. PMID: 21545258 Free PMC article.
-
Pathogen chip for respiratory tract infections.J Clin Microbiol. 2013 Mar;51(3):945-53. doi: 10.1128/JCM.02317-12. Epub 2013 Jan 9. J Clin Microbiol. 2013. PMID: 23303493 Free PMC article.
-
Large-scale evolutionary surveillance of the 2009 H1N1 influenza A virus using resequencing arrays.Nucleic Acids Res. 2010 May;38(9):e111. doi: 10.1093/nar/gkq089. Epub 2010 Feb 25. Nucleic Acids Res. 2010. PMID: 20185568 Free PMC article.
References
-
- Mehlmann M, Dawson ED, Townsend MB, Smagala JA, Moore CA, Smith CB, Cox NJ, Kuchta RD, Rowlen KL. Robust sequence selection method used to develop the FluChip diagnostic microarray for influenza virus. Journal of Clinical Microbiology. 2006;44:2857–2862. doi: 10.1128/JCM.00135-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

