Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana
- PMID: 18783613
- PMCID: PMC2551617
- DOI: 10.1186/1471-2229-8-92
Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana
Abstract
Background: DNA methylation is an important biochemical mark that silences repetitive sequences, such as transposons, and reinforces epigenetic gene expression states. An important class of repetitive genes under epigenetic control in eukaryotic genomes encodes ribosomal RNA (rRNA) transcripts. The ribosomal genes coding for the 45S rRNA precursor of the three largest eukaryotic ribosomal RNAs (18S, 5.8S, and 25-28S) are found in nucleolus organizer regions (NORs), comprised of hundreds to thousands of repeats, only some of which are expressed in any given cell. An epigenetic switch, mediated by DNA methylation and histone modification, turns rRNA genes on and off. However, little is known about the mechanisms that specify and maintain the patterns of NOR DNA methylation.
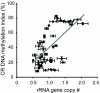
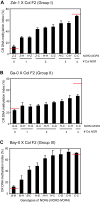
Results: Here, we explored the extent of naturally-occurring variation in NOR DNA methylation among accessions of the flowering plant Arabidopsis thaliana. DNA methylation in coding regions of rRNA genes was positively correlated with copy number of 45S rRNA gene and DNA methylation in the intergenic spacer regions. We investigated the inheritance of NOR DNA methylation patterns in natural accessions with hypomethylated NORs in inter-strain crosses and defined three different categories of inheritance in F1 hybrids. In addition, subsequent analysis of F2 segregation for NOR DNA methylation patterns uncovered different patterns of inheritance. We also revealed that NOR DNA methylation in the Arabidopsis accession Bor-4 is influenced by the vim1-1 (variant in methylation 1-1) mutation, but the primary effect is specified by the NORs themselves.
Conclusion: Our results indicate that the NORs themselves are the most significant determinants of natural variation in NOR DNA methylation. However, the inheritance of NOR DNA methylation suggests the operation of a diverse set of mechanisms, including inheritance of parental methylation patterns, reconfiguration of parental NOR DNA methylation, and the involvement of trans-acting modifiers.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

