Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods
- PMID: 18783629
- PMCID: PMC2547107
- DOI: 10.1186/1472-6750-8-69
Measuring microRNAs: comparisons of microarray and quantitative PCR measurements, and of different total RNA prep methods
Abstract
Background: Determining the expression levels of microRNAs (miRNAs) is of great interest to researchers in many areas of biology, given the significant roles these molecules play in cellular regulation. Two common methods for measuring miRNAs in a total RNA sample are microarrays and quantitative RT-PCR (qPCR). To understand the results of studies that use these two different techniques to measure miRNAs, it is important to understand how well the results of these two analysis methods correlate. Since both methods use total RNA as a starting material, it is also critical to understand how measurement of miRNAs might be affected by the particular method of total RNA preparation used.
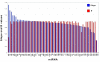
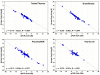
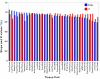
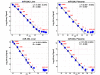
Results: We measured the expression of 470 human miRNAs in nine human tissues using Agilent microarrays, and compared these results to qPCR profiles of 61 miRNAs in the same tissues. Most expressed miRNAs (53/60) correlated well (R > 0.9) between the two methods. Using spiked-in synthetic miRNAs, we further examined the two miRNAs with the lowest correlations, and found the differences cannot be attributed to differential sensitivity of the two methods. We also tested three widely-used total RNA sample prep methods using miRNA microarrays. We found that while almost all miRNA levels correspond between the three methods, there were a few miRNAs whose levels consistently differed between the different prep techniques when measured by microarray analysis. These differences were corroborated by qPCR measurements.
Conclusion: The correlations between Agilent miRNA microarray results and qPCR results are generally excellent, as are the correlations between different total RNA prep methods. However, there are a few miRNAs whose levels do not correlate between the microarray and qPCR measurements, or between different sample prep methods. Researchers should therefore take care when comparing results obtained using different analysis or sample preparation methods.
Figures




Similar articles
-
Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis.BMC Genomics. 2009 Aug 28;10:407. doi: 10.1186/1471-2164-10-407. BMC Genomics. 2009. PMID: 19715577 Free PMC article.
-
Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs.Biotechniques. 2010 Mar;48(3):219-22. doi: 10.2144/000113367. Biotechniques. 2010. PMID: 20359303
-
Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression.RNA. 2010 May;16(5):991-1006. doi: 10.1261/rna.1947110. Epub 2010 Apr 1. RNA. 2010. PMID: 20360395 Free PMC article.
-
Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available.Methods. 2010 Apr;50(4):244-9. doi: 10.1016/j.ymeth.2010.01.026. Epub 2010 Jan 28. Methods. 2010. PMID: 20109550 Review.
-
Multiplexed detection methods for profiling microRNA expression in biological samples.Angew Chem Int Ed Engl. 2008;47(4):644-52. doi: 10.1002/anie.200702450. Angew Chem Int Ed Engl. 2008. PMID: 17994653 Free PMC article. Review.
Cited by
-
Strand displacement-triggered G-quadruplex/rolling circle amplification strategy for the ultra-sensitive electrochemical sensing of exosomal microRNAs.Mikrochim Acta. 2020 Feb 15;187(3):172. doi: 10.1007/s00604-020-4143-9. Mikrochim Acta. 2020. PMID: 32062754
-
Methodological challenges in utilizing miRNAs as circulating biomarkers.J Cell Mol Med. 2014 Mar;18(3):371-90. doi: 10.1111/jcmm.12236. Epub 2014 Feb 18. J Cell Mol Med. 2014. PMID: 24533657 Free PMC article. Review.
-
Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal.BMC Biotechnol. 2018 Mar 16;18(1):16. doi: 10.1186/s12896-018-0421-6. BMC Biotechnol. 2018. PMID: 29548320 Free PMC article.
-
Influence of RNA labeling on expression profiling of microRNAs.J Mol Diagn. 2012 Jan;14(1):12-21. doi: 10.1016/j.jmoldx.2011.08.005. Epub 2011 Nov 7. J Mol Diagn. 2012. PMID: 22074760 Free PMC article.
-
Performance comparison of digital microRNA profiling technologies applied on human breast cancer cell lines.PLoS One. 2013 Oct 8;8(10):e75813. doi: 10.1371/journal.pone.0075813. eCollection 2013. PLoS One. 2013. PMID: 24116077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

