F-Seq: a feature density estimator for high-throughput sequence tags
- PMID: 18784119
- PMCID: PMC2732284
- DOI: 10.1093/bioinformatics/btn480
F-Seq: a feature density estimator for high-throughput sequence tags
Abstract
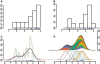
Tag sequencing using high-throughput sequencing technologies are now regularly employed to identify specific sequence features, such as transcription factor binding sites (ChIP-seq) or regions of open chromatin (DNase-seq). To intuitively summarize and display individual sequence data as an accurate and interpretable signal, we developed F-Seq, a software package that generates a continuous tag sequence density estimation allowing identification of biologically meaningful sites whose output can be displayed directly in the UCSC Genome Browser.
Availability: The software is written in the Java language and is available on all major computing platforms for download at http://www.genome.duke.edu/labs/furey/software/fseq.
Figures
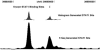
References
-
- Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. - PubMed
-
- Parzen E. On the estimation of a probability density function and mode. Ann. Math. Stat. 1962;33:1065–1076.
-
- Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

